Article highlights & insights
Many primary-tumour subregions have low levels of molecular oxygen, termed hypoxia. Hypoxic tumours are at elevated risk for local failure and distant metastasis, but the molecular hallmarks of tumour hypoxia remain poorly defined.
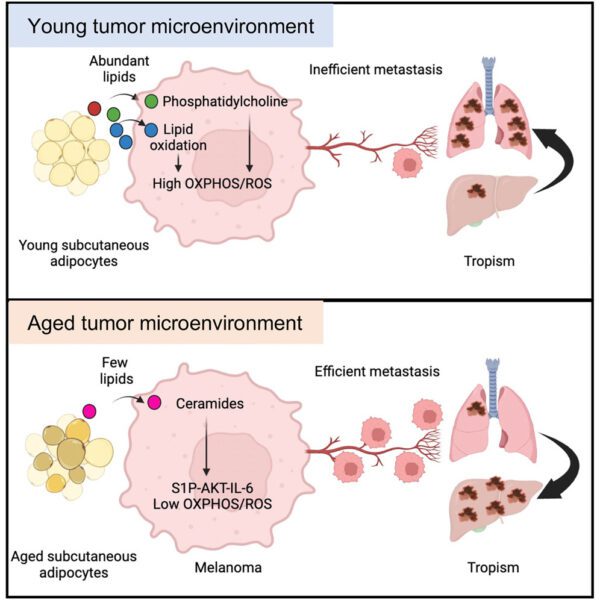
To fill this gap, we quantified hypoxia in 8,006 tumours across 19 tumour types. In ten tumour types, hypoxia was associated with elevated genomic instability. In all 19 tumour types, hypoxic tumours exhibited characteristic driver-mutation signatures. We observed widespread hypoxia-associated dysregulation of microRNAs (miRNAs) across cancers and functionally validated miR-133a-3p as a hypoxia-modulated miRNA. In localised prostate cancer, hypoxia was associated with elevated rates of chromothripsis, allelic loss of PTEN and shorter telomeres. These associations are particularly enriched in polyclonal tumors, representing a constellation of features resembling tumour nimbosus, an aggressive cellular phenotype.
Overall, this work establishes that tumour hypoxia may drive aggressive molecular features across cancers and shape the clinical trajectory of individual tumours.
Many primary-tumour subregions have low levels of molecular oxygen, termed hypoxia. Hypoxic tumours are at elevated risk for local failure and distant metastasis, but the molecular hallmarks of tumour hypoxia remain poorly defined.
To fill this gap, we quantified hypoxia in 8,006 tumours across 19 tumour types. In ten tumour types, hypoxia was associated with elevated genomic instability. In all 19 tumour types, hypoxic tumours exhibited characteristic driver-mutation signatures. We observed widespread hypoxia-associated dysregulation of microRNAs (miRNAs) across cancers and functionally validated miR-133a-3p as a hypoxia-modulated miRNA. In localised prostate cancer, hypoxia was associated with elevated rates of chromothripsis, allelic loss of PTEN and shorter telomeres. These associations are particularly enriched in polyclonal tumors, representing a constellation of features resembling tumour nimbosus, an aggressive cellular phenotype.
Overall, this work establishes that tumour hypoxia may drive aggressive molecular features across cancers and shape the clinical trajectory of individual tumours.
Institute Authors
Groups
Group leader
Research topics & keywords
Meet the Research Team
All publications
https://doi.org/10.1038/s44161-025-00740-z
Single-cell profiling reveals three endothelial-to-hematopoietic transitions with divergent isoform expression landscapes
11 November 2025
Institute Authors (6)
Robert Sellers, John Weightman, Wolfgang Breitwieser, Natalia Moncaut, Michael Lie-a-ling, Georges Lacaud
Labs & Facilities
Computational Biology Support, Molecular Biology, Genome Editing and Mouse Models
Research Group
Stem Cell Biology
11 November 2025
https://doi.org/10.1136/jitc-2025-012527
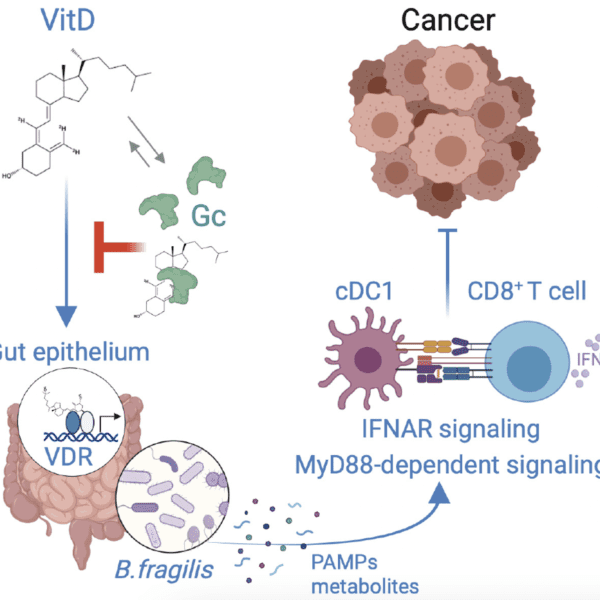
Systemic immunosuppression from ultraviolet radiation exposure inhibits cancer immunotherapy
31 October 2025
Institute Authors (4)
Isabella Mataloni, Antonia Banyard, Garry Ashton, Amaya Virós
Labs & Facilities
Mass and Flow Cytometry, Histology
Research Group
Skin Cancer & Ageing
31 October 2025
https://aacrjournals.org/cancerdiscovery/article/doi/10.1158/2159-8290.CD-24-1224/766638/Glucocorticoids-Unleash-Immune-dependent-Melanoma
Glucocorticoids Unleash Immune-dependent Melanoma Control through Inhibition of the GARP/TGF β Axis
15 October 2025
Institute Authors (12)
Charles Earnshaw, Poppy Dunn, Shih-Chieh Chiang, Maria Koufaki, Massimo Russo, Kimberley Hockenhull, Erin Richardson, Anna Pidoux, Alex Baker, Richard Reeves, Robert Sellers, Sudhakar Sahoo
Labs & Facilities
Computational Biology Support, Visualisation, Irradiation and Analysis
Research Group
Cancer Inflammation and Immunity
15 October 2025
/wp-content/uploads/2025/09/Annual_Report_2024.pdf
2024 Annual Report
23 September 2025
23 September 2025
https://doi.org/10.1182/blood.2024028033
An in vivo barcoded CRISPR-Cas9 screen identifies Ncoa4-mediated ferritinophagy as a dependence in Tet2-deficient hematopoiesis
4 September 2025
Institute Authors (1)
Justin Loke
Research Group
Myeloid Cancer Biology
4 September 2025
https://doi.org/10.1038/s41467-024-49692-1
Whole genome sequencing refines stratification and therapy of patients with clear cell renal cell carcinoma
15 July 2025
Institute Authors (1)
Samra Turajlić
Research Group
Cancer Dynamics
15 July 2025
Our Research
Our research spans the whole spectrum of cancer research from cell biology through to translational and clinical studies
Research Groups
Our research groups study many fundamental questions of cancer biology and treatment
Our Facilities
The Institute has outstanding core facilities that offer cutting edge instruments and tailored services from expert staff
Latest News & Updates
Find out all our latest news