The Scientific Computing core facility (SciCom) supports translational cancer research by providing storage and compute services for scientists and core facilities at the Cancer Research UK Manchester Institute.
Our goal is to find and provide the computing tools and resources our scientists need to carry out their outstanding cancer research. For this, SciCom operates a highly integrated data analysis platform, consisting of a High-Performance-Computing system, a Linux virtualisation platform, bare metal servers and cloud services.
The IT and Scientific Computing core facility supports translational cancer research by providing essential IT services, large research IT storage and computing facilities for scientists, and core facilities at the Cancer Research UK Manchester Institute. Our mission is to provide practical and efficient solutions for our users to cover the entire data analysis lifecycle.

The key services provided by the IT and Scientific Computing core facility are:
- High Performance Computing service including support
- Solution for Virtual Desktop Infrastructure (Linux/Windows) for large scale data analysis
- Storage facilities and consultancy for business data and large-scale research data
- Software development support for business and research application and pipelines
- Computer, IT installation and user support & management
Griffin High Performance Computing Cluster
- 100 x standard nodes, 48 CPU cores, 256GB RAM
- 2 x 4 TB High Memory nodes, 96 CPU cores, 4096GB RAM
- 1 x NVIDIA Redstone system, 48 CPU cores, 512GB RAM, 4 x NVIDIA A100 connected via NVLINK
- HDR 100Gb/s InfiniBand Network
- 1PB Panasas Active Store Parallel File System
High Performance Virtualisation Platform
- 7 oVirt Hypervisor nodes
- 21TB Hybrid storage for virtual drives
3.1 PB IBM Spectrum Scale based research storage for large datasets
2x 400TB High Availability Windows File Storage
2x mirrored VMware based virtualisation platform for business critical Virtual Machines
Featured Publications
2023 Annual Report
13th September 2024
Our Aims
We aim to find and provide the computing tools and resources our scientists need to carry out their outstanding cancer research. For this, the facility operates a highly integrated data analysis platform, consisting of a High-Performance-Computing system, a Linux virtualisation platform, bare metal servers and cloud services. The platform provides solutions for data storage, including automated data management tools, as well as data processing, analysis, publication, and archiving.
It allows the secure processing and analysis of sensitive high-throughput data. In addition, we offer application and software development support to enable our scientists to use the latest Bioinformatics methods and technologies for their research.
Close collaborations
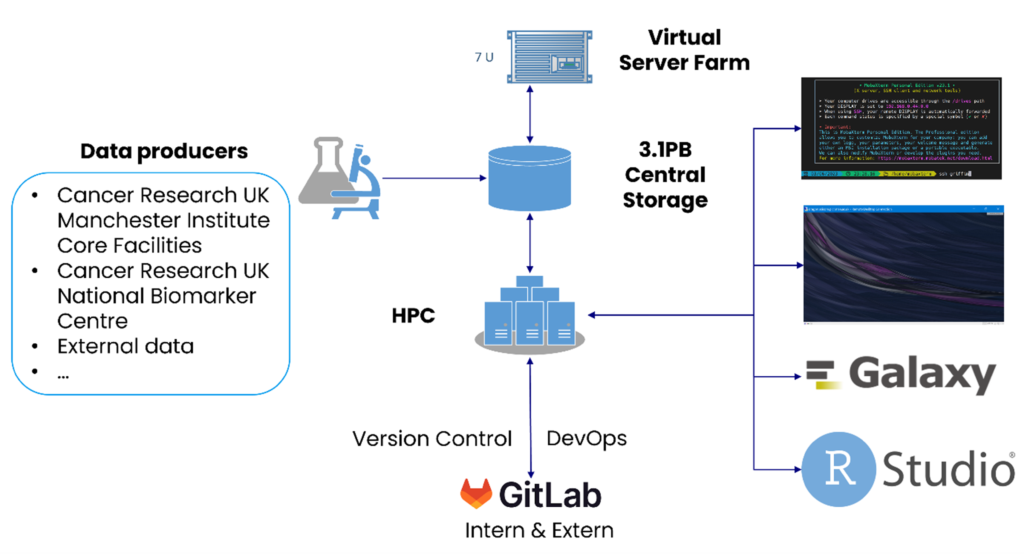
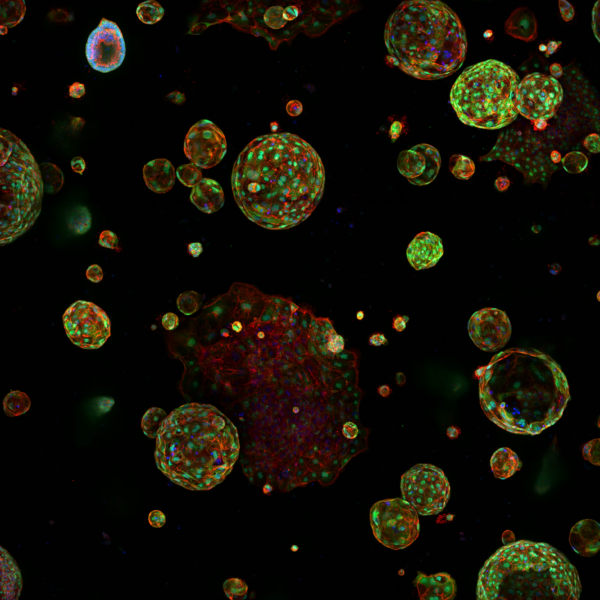
The Institute operates a multitude of cutting edge instruments for conducting genomics, proteomic and imaging experiments. The large amounts of data produced by these instruments are stored on a 4PB storage system and can be analysed using an oVirt-based virtualisation platform or “Griffin”. Griffin is a heterogenous 1,500 core High Performance Compute Linux cluster, consisting of standard, high memory and GPU nodes, with a 1PB parallel file system. With its tightly integrated hardware and cloud infrastructure, SciCom operates Linux and Windows-based high throughput data analysis and management services.
We work in partnership with the Computational Biology Support Team – find out more about their work on their Facility page.

Virtual machines, practical support
Combining virtual machines hosted on powerful computing hardware with remote visualisation also allows interactive data processing of compute, data, and memory intensive workloads, which is especially helpful for processing proteomics data. Special data protection arrangements on Griffin allows the processing and analysis of access-controlled (e.g. dbGaP, ICGC, etc.) and clinical trial data.
Scientific Computing has a strong focus on automating the processing of data to increase throughput and accelerate and ease the burden of data analysis for the scientists.
Griffin
Griffin is a heterogeneous InfiniBand Linux cluster designed to analyse a large variety of different data types using different methods and algorithms. It consists of 100 x standard compute nodes, 2 x high memory nodes, and an NVIDIA Redstone GPU (graphic processing units) system and an FPGA. We decided to replace the former MOAB/Torque-based batch system with the modern and widely used SLURM for more effective job management. HPC nodes and storage are connected via a high-speed InfiniBand 100Gb/s connection, allowing high-speed data transfer between the components. Accessing the system is also much faster than before, as the network bandwidth has been upgraded from 10GbE to 25GbE.
With its massive computing power, Griffin is a crucial component of SciCom’s High Throughput Data Analysis platform. It is tightly integrated with various storage systems, the High-Performance virtualisation platform, bare metal servers and cloud services to provide an integrated platform for analysing research and clinical data. The platform covers the entire data analysis lifecycle, spanning from data generation, processing, downstream analysis, and visualisation, publication, and archiving, while following FAIR principles.

Meet the IT and Scientific Computing team
Our IT and Scientific Computing team cover a wide range of activities which are essential for the running of much of the research across the Institute, as well as collaborations between diverse teams. Together they:
- Run the IT helpdesk and respond to requests from staff and students, including purchasing, maintaining and repairing computer hardware and software
- Oversee the digital architecture of our scientific storage infrastructure, supporting research that involves managing extremely large datasets and complex integrations between different software pipelines and workflows
- Support other IT systems including web design and our intranet
- Manage the whole team, thinking strategically about new technologies, software and data security applications
Get in touch
Our vision for world leading cancer research in the heart of Manchester
We are a leading cancer research institute within The University of Manchester, spanning the whole spectrum of cancer research – from investigating the molecular and cellular basis of cancer, to translational research and the development of therapeutics.
Our collaborations
Bringing together internationally renowned scientists and clinicians
Scientific Advisory Board
Supported by an international Scientific Advisory Board
Careers that have a lasting impact on cancer research and patient care
We are always on the lookout for talented and motivated people to join us. Whether your background is in biological or chemical sciences, mathematics or finance, computer science or logistics, use the links below to see roles across the Institute in our core facilities, operations teams, research groups, and studentships within our exceptional graduate programme.
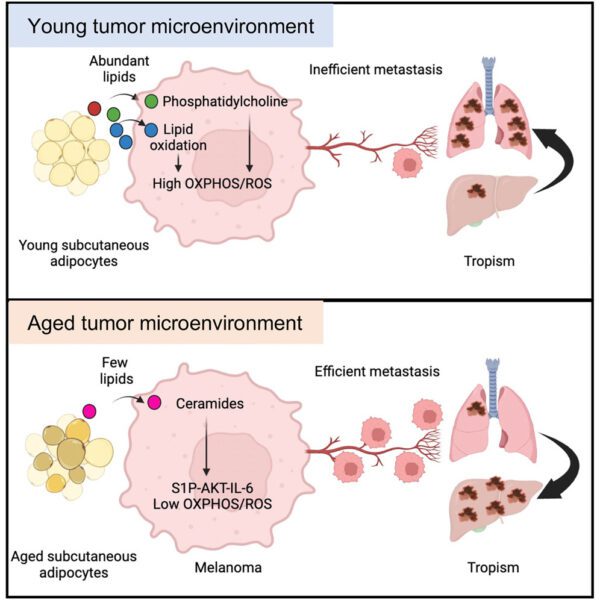
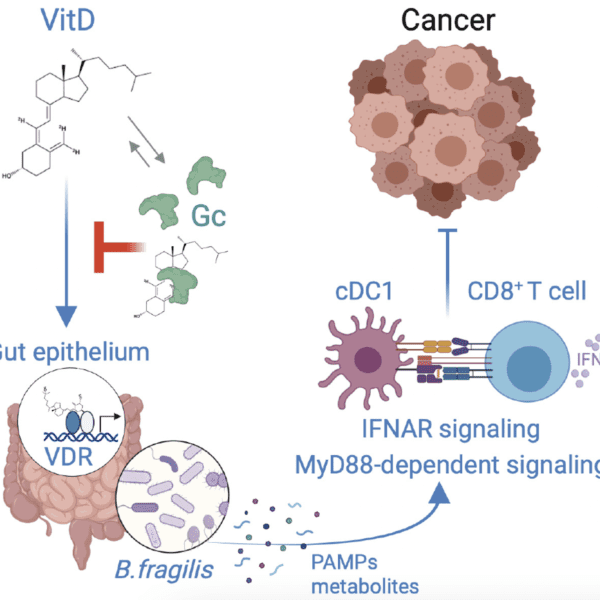


A note from the Team Leader – Marek Dynowski
I am dedicated to managing a top-tier IT and Scientific Computing Core Facility, ensuring high user satisfaction and equipping researchers with the essential tools to advance their work. My primary focus is integrating cybersecurity standards within IT and Scientific Computing, despite their differing priorities. Additionally, I emphasise utilising software development methodologies, such as CI/CD, to automate IT administration and streamline Bioinformatic Pipeline development on HPC systems.