The Molecular Biology Core provides a wide range of services for precision and high throughput analysis of DNA and RNA to assist in molecular biology projects within the CRUK Manchester Institute. We have developed a multitude of workflows for automated assay running and support scientists with state-of-the-art analytical instrumentation.
We support a wide range of key services for research across the Institute, including:
- Clonal Sequencing
- Cell line authentication
- Compound Screening
- Long read sequencing
- Pathogen screening
- Single Cell transcriptomics
- Spatial transcriptomics
- Sample QC
Featured Publications
2023 Annual Report
13th September 2024
Long Read Sequencing
Major advances in throughput and data accuracy have led to a significant expansion of nanopore based long-read sequencing applications. The service currently employs Oxford Nanopore Technology (ONT) MinIon and PromethION sequencers for a variety of methodologies, including whole genome sequencing, methylation calling, cfDNA, single cell and direct RNA sequencing, as well as metagenomic analysis.

Single Cell Technologies
Most single cell sequencing projects are processed using 10X Genomics emulsion-based technology, including scRNA-Seq, immune cell profiling, multiome (scRNA & ATAC-Seq), and CRISPR screen projects (e.g. Perturb-Seq). An additional frequent application is Smart-Seq for single cell transcriptome profiling of rare cell populations. For this microwell plate-based technology the service has adopted a miniaturisation/automation approach, employing the department’s robotic platforms, including Bravo (Agilent), Mantis (Formulatrix) and Echo 550 (Beckman). Furthermore, automation solutions are currently explored for additional single cell technologies, including Combinatorial Indexing.
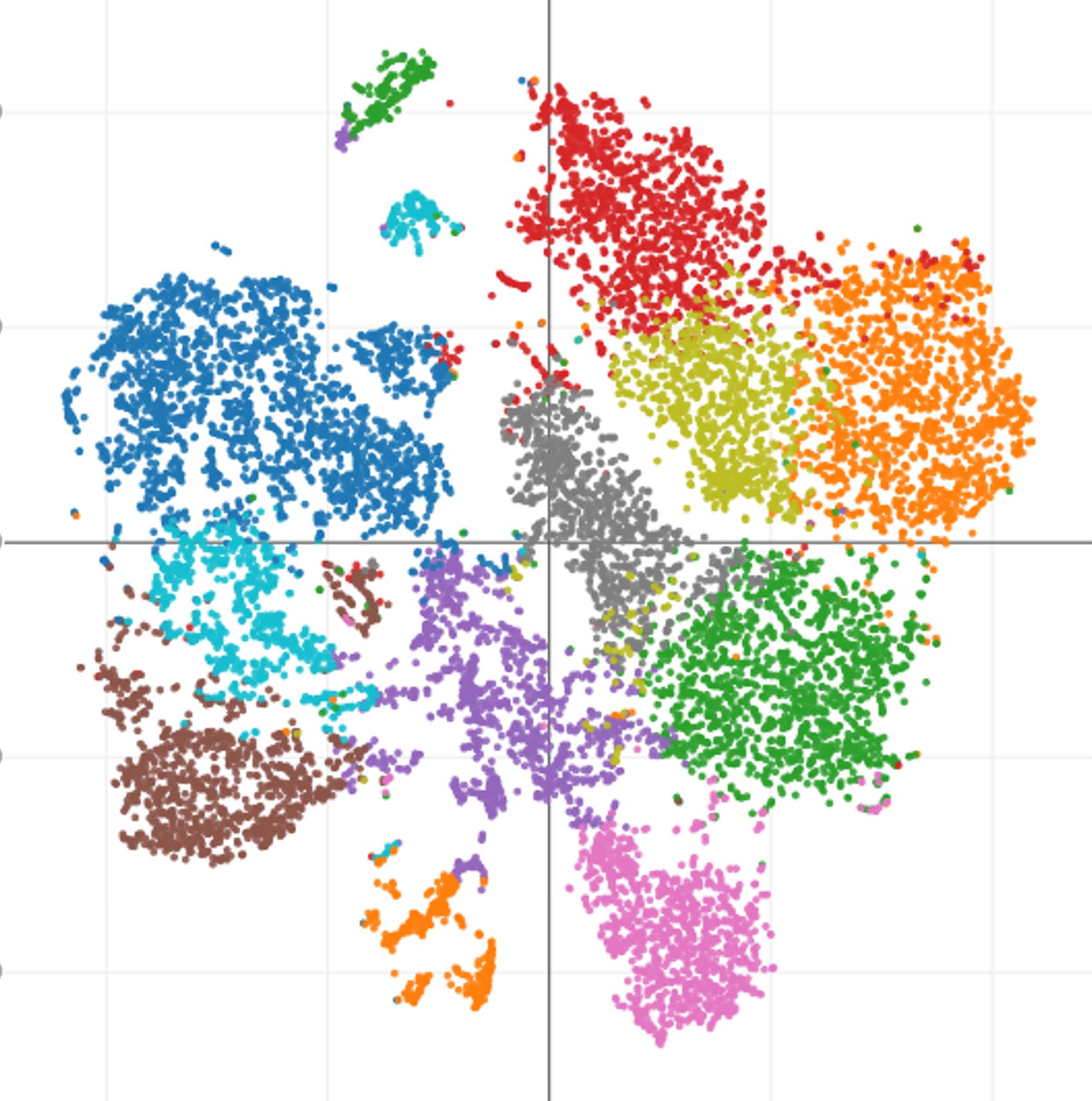
Clonal Sequencing
This comprehensive service covers all aspects of Next Generation Sequencing (NGS). An extensive portfolio of library prep methods for RNA and DNA sequencing is aimed at coping with a wide range in quantity and quality of inputs. Most NGS projects are processed in automated workflows using high throughput instrumentation, e.g. the Agilent Bravo liquid handling robot. The bulk of sequencing is performed on Illumina instruments, predominantly the high throughput platform, Novaseq 6000.

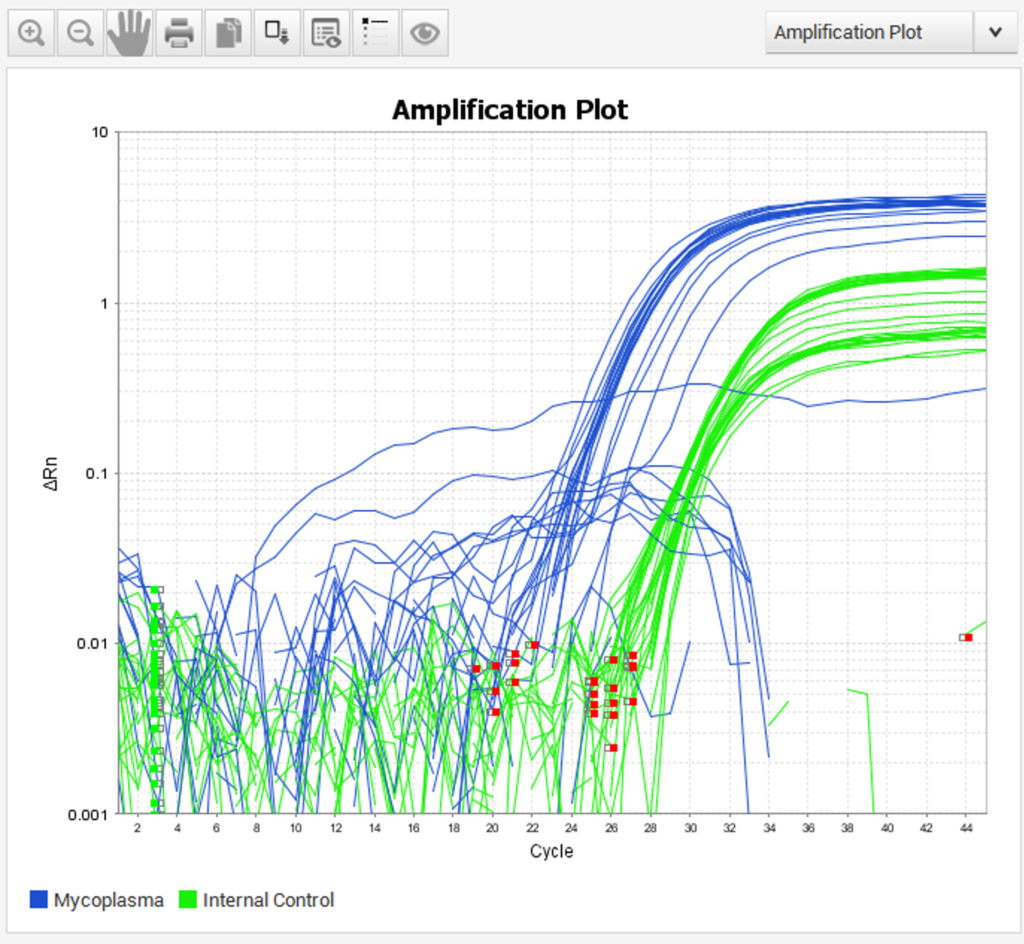
Pathogen Screening
To help in compliance with the Institute’s policy on Mycoplasma control, we offer a regular screening service for Mycoplasma infection of cell lines. In addition, prior to injection into mice relevant cell line samples are screened for Mycoplasma and Murine Hepatitis Virus (MHV).
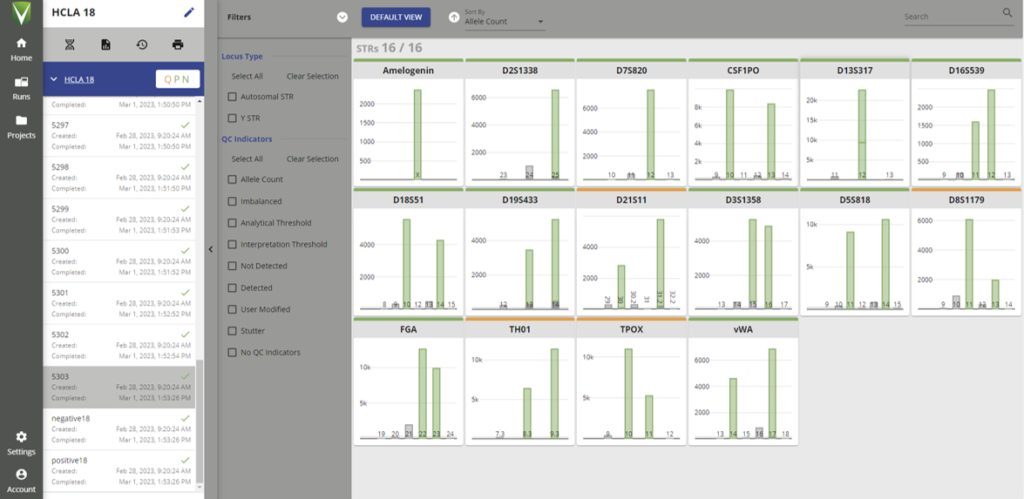
Human Cell Line Authentication
To maintain the highest standards of experimental work CRUK MI encourages regular testing of cultured cell lines for their authenticity. For accurate identification we offer a NGS based assay which detects highly variable short tandem repeat (STR) regions in the genome. Test results are cross-checked against an extensive in-house cell line database as well as ATCC references.
Sample QC
Many Molecular Biology applications, e.g. Next Generation Sequencing, require a minimum quality of their input material. To ascertain this, we offer a QC service for extracted DNA and RNA by processing sample aliquots on dedicated instrumentation, including Qubit (Thermo Fisher) for quantitation, as well as Bioanalyser and Fragment Analyser (Agilent) for quality analysis.

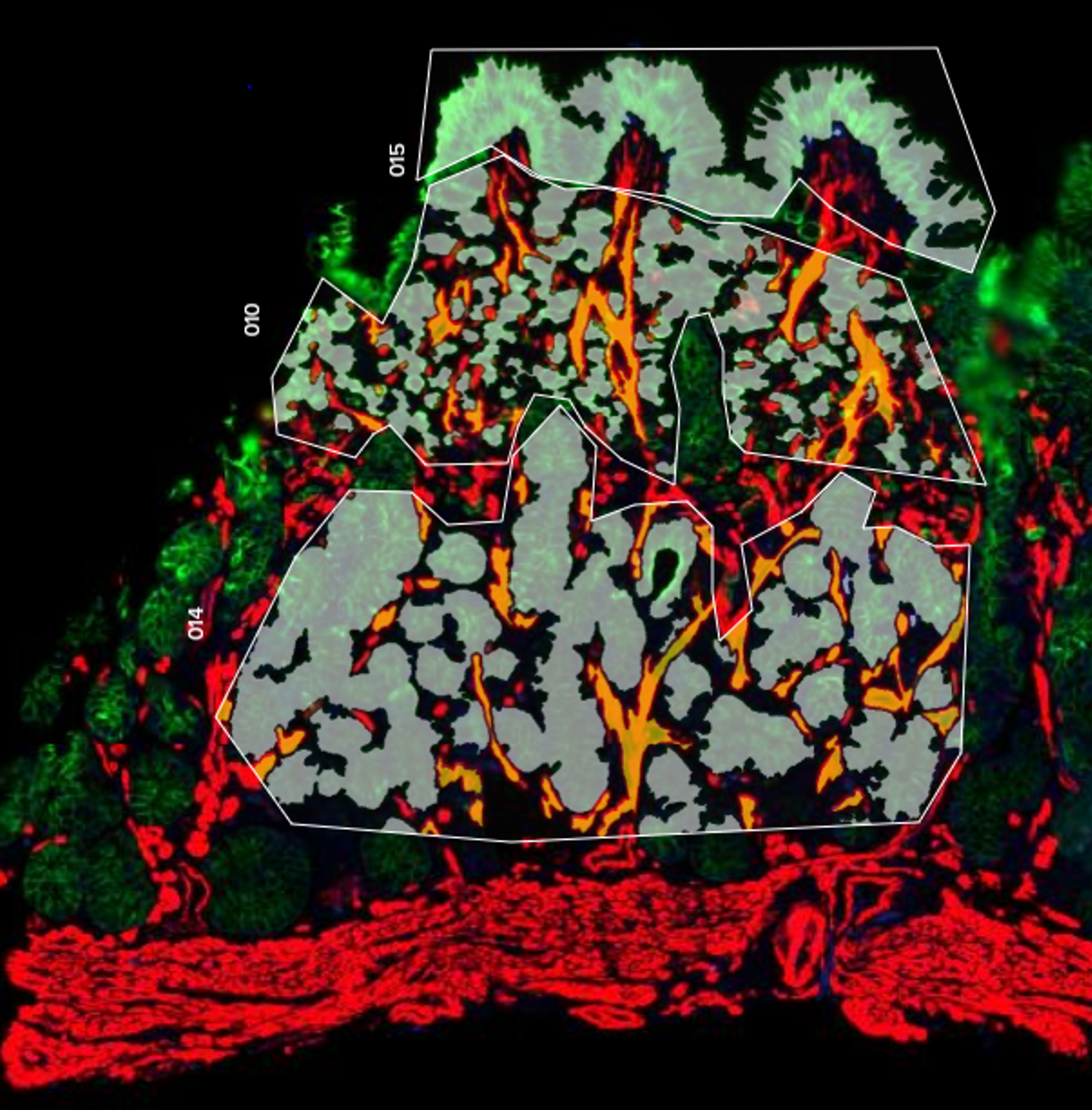
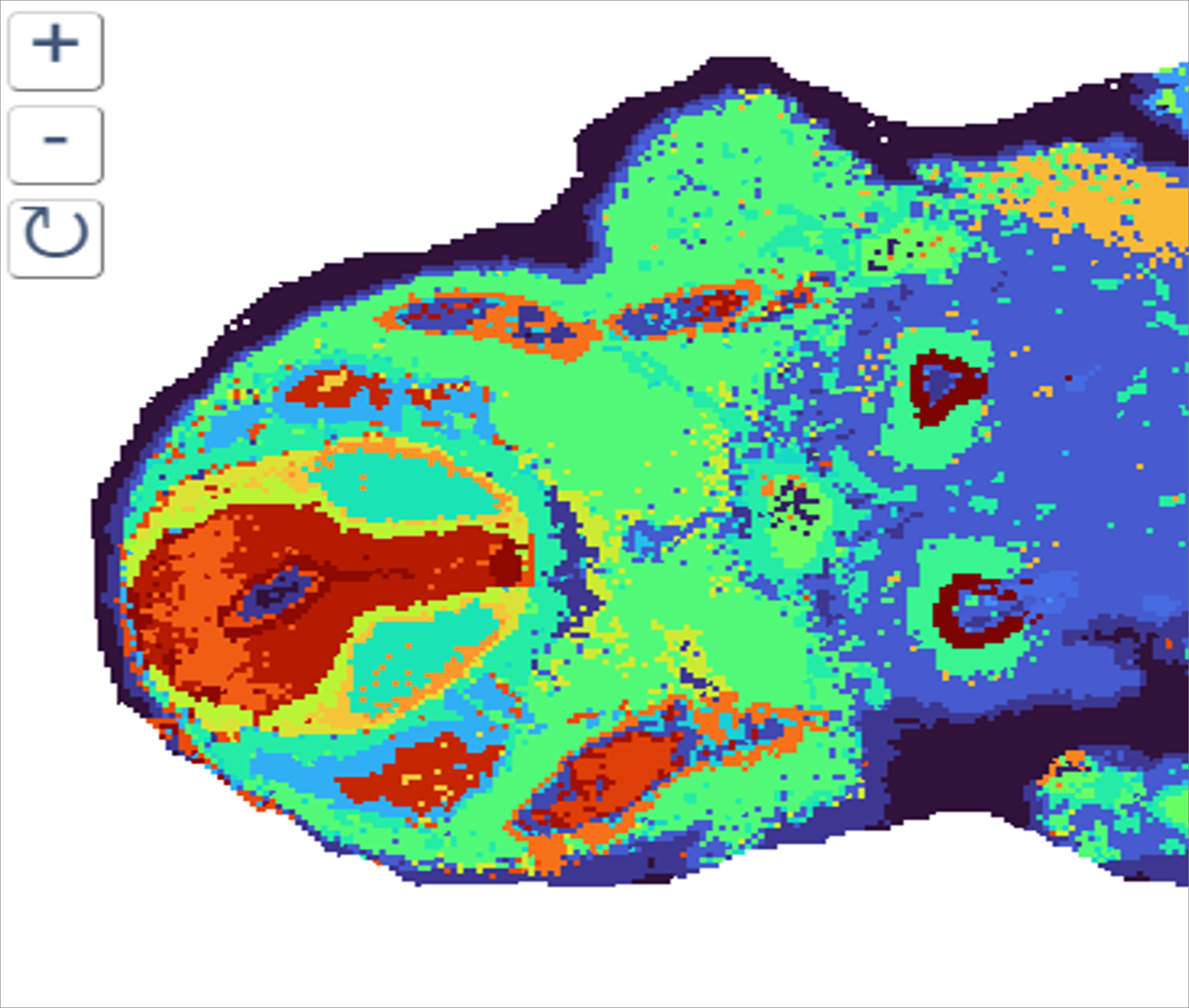
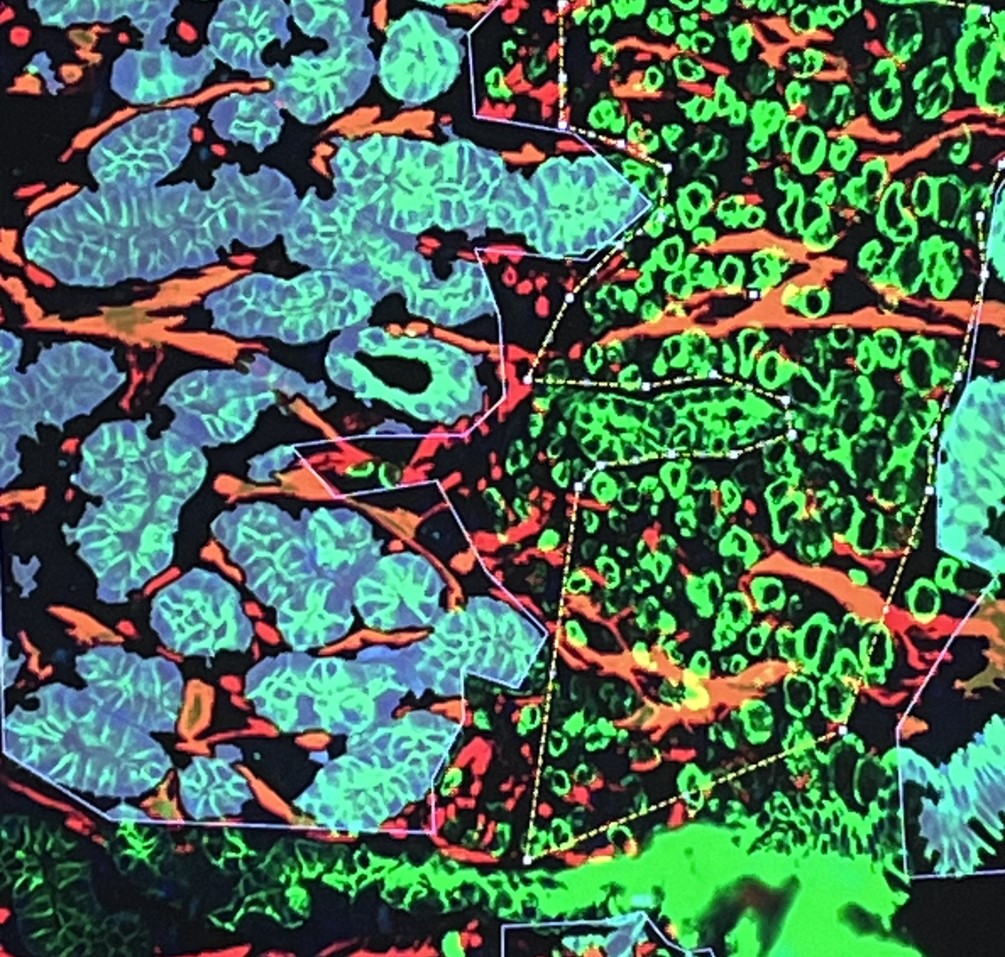
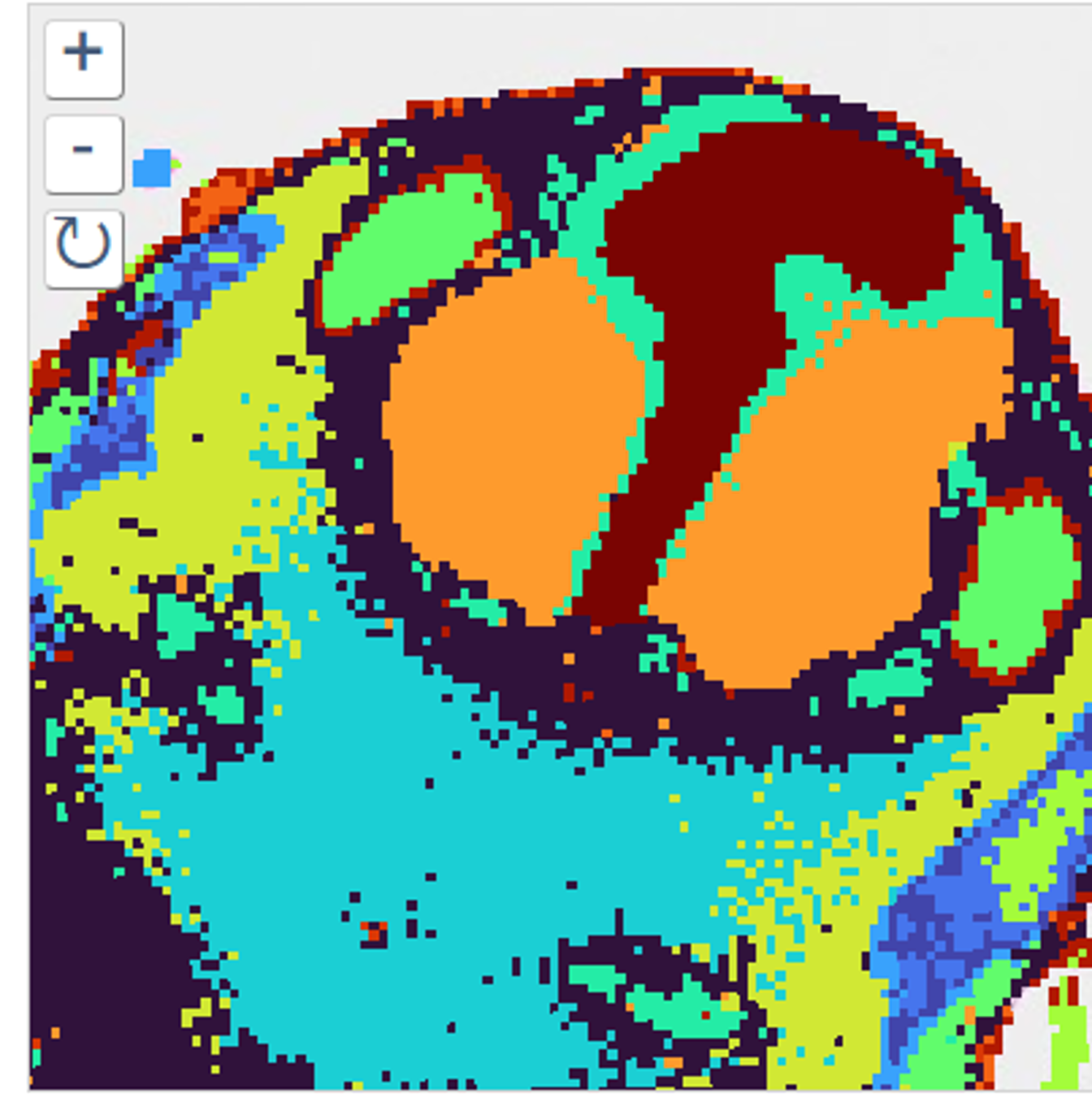
Spatial Transcriptomics/Proteomics
Sequencing based spatial omics is currently supported by three technology areas, 10X Genomics Visium, STOmics Stereo-seq, and Nanostring GeoMx, and is a multi-Core Facility spanning service.
The service is comprehensive and includes all aspects of tissue processing (by Histology), tissue imaging (VIA), region selection by the Service Users, followed by probe capture, library prep and sequencing by Molecular Biology. Read conversions and QC pipelines are administered by SciCom and CBS.
Get in touch
Our vision for world leading cancer research in the heart of Manchester
We are a leading cancer research institute within The University of Manchester, spanning the whole spectrum of cancer research – from investigating the molecular and cellular basis of cancer, to translational research and the development of therapeutics.
Our collaborations
Bringing together internationally renowned scientists and clinicians
Scientific Advisory Board
Supported by an international Scientific Advisory Board
Careers that have a lasting impact on cancer research and patient care
We are always on the lookout for talented and motivated people to join us. Whether your background is in biological or chemical sciences, mathematics or finance, computer science or logistics, use the links below to see roles across the Institute in our core facilities, operations teams, research groups, and studentships within our exceptional graduate programme.
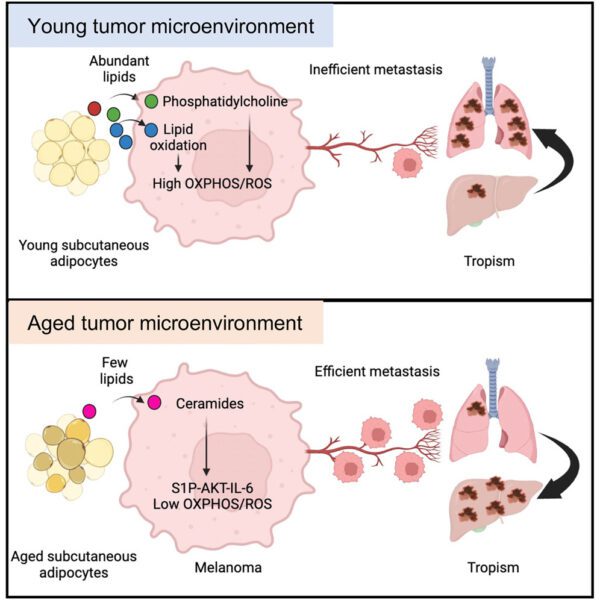
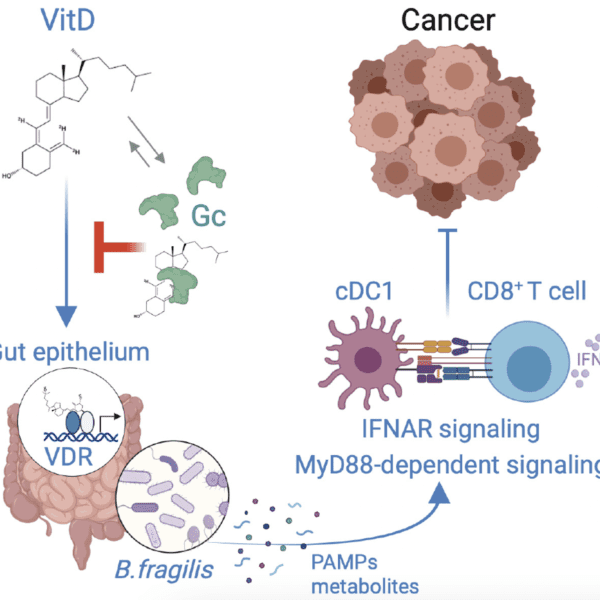





A note from the Team Leader – Wolfgang Breitwieser
With the capabilities to offer a diverse range of technologies – including spatial transcriptomics – the Institute Core teams are now in a unique position to offer tailored solutions to individual scientific challenges in this field.