The Visualisation, Irradiation and Analysis (VIA) facility within Cancer Research UK Manchester Institute has a large remit covering the training of researchers, operation of instruments, supply of training & support, maintenance, quality control of hardware and outputs, and experimental design for:
- Confocal and widefield-based high content screening (HCS).
- Whole-slide histology imaging (brightfield, polarised, fluorescence and hi-plex).
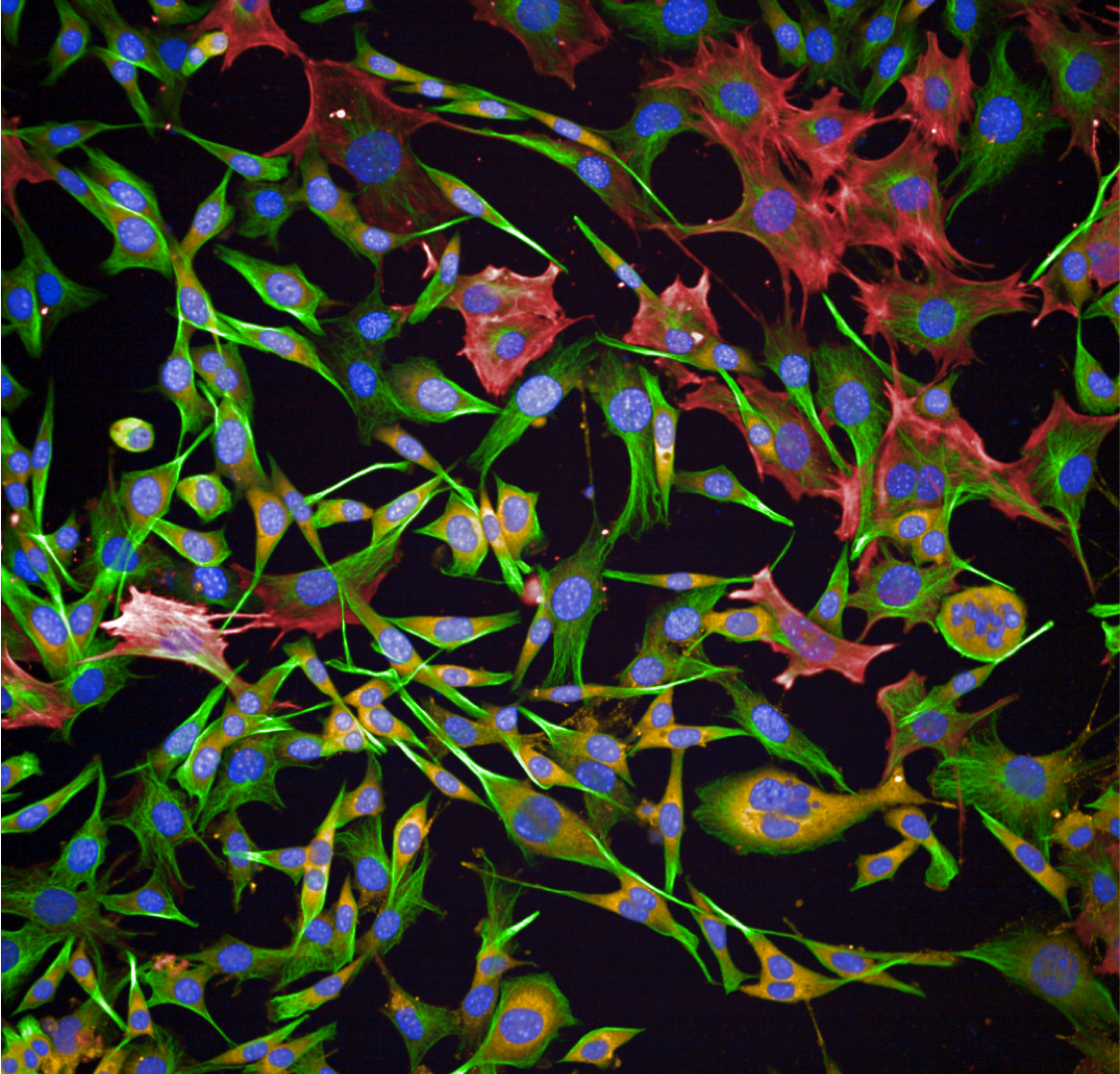
- Microscopy (widefield, laser scanning confocal, spinning disk confocal, two-photon, super-resolution).
- Image processing and analysis support, commercial & open software.
- MI & OCRB Ionised and non-ionised irradiation (monitoring, safety & dosimetry).
- Maintenance and support of research lab-based imaging equipment
The Facility supports researchers by supplying maintained instruments, and increases their knowledge by providing training, so that researcher operating the equipment have the necessary experience and familiarity of the biological samples and understand the relevancy of the systems and processes.
The workload is split between projects that are imaging only and those that are multi-facility based. The team within the facility are on hand to offer training, support and assist in the operation of the instrument if required.
Instruments are offered on a facility or service level; however, some modalities are completed by the facility team to enable high throughput of mixed samples from multiple researchers, Automated whole slide imaging systems with brightfield, fluorescence (penta-Semock or Kromnigon filter sets.
- Evident VS120, IHC and fluorescence whole slide imager, automation for 120 slides.
- Evident VS200, IHC, polarised and fluorescence whole slide imager, automation for 200 slides (standard & mega).
- Akoya Phenocycler Fusion II, dual imaging of tissue sections utilising up to 99 fluorescent labels by cyclic imaging.
We provide a range of instruments which researchers can be trained to utilise directly, or where one of our team can assist in image capture. All instruments undergo a range of quality control measures to ensure data integrity.
- Incucyte S3. Incubator based imaging of up to six vessels (petri, multi-well plate)
- Incucyte S5. Incubator based imaging of up to six vessels (petri, multi-well plate)
- Leica TCS confocal and 2photon. Upright microscope for thick tissue imaging
- Revvity Opera Phenix, confocal high content screening with sample automation.
- Revvity Operetta, widefield high content screening.
- Zeiss LSM880 Airyscan, confocal and super resolution for fixed live cell imaging.
- Workstation and server-based support for open-source and commercial software. Support for, amongst others, Cellprofiller, ImageJ, Napari, QuPath, Halo & Huygens.
- Virtual PCs hosting an array of open source software for microscopy, tissue, pre-clinical and high content imaging
- Ultivue S5 3D printer.
More instruments coming soon …
Featured Publications
2023 Annual Report
13th September 2024
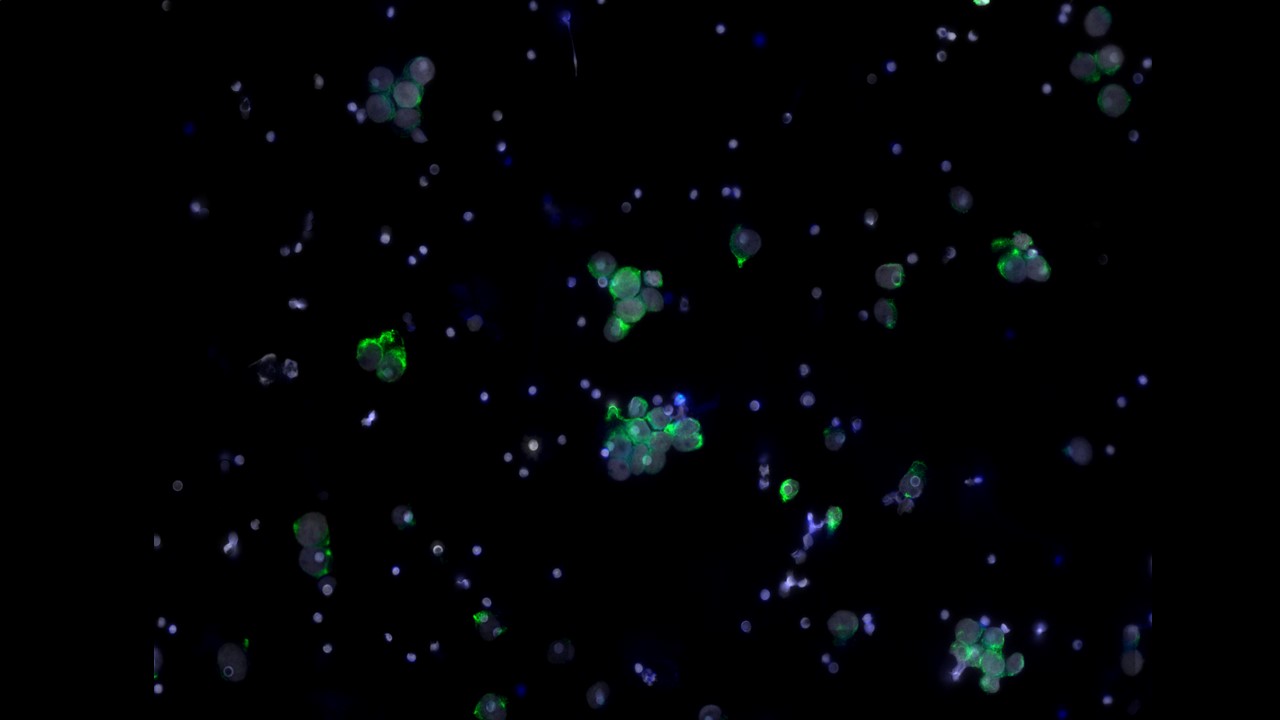
High Content Screening
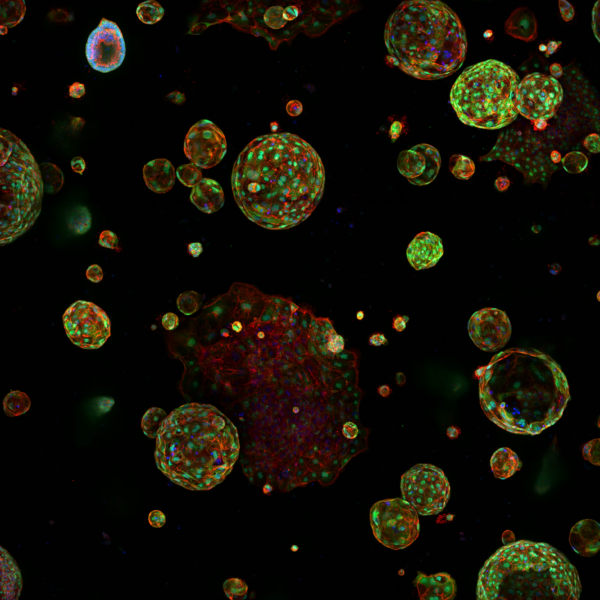
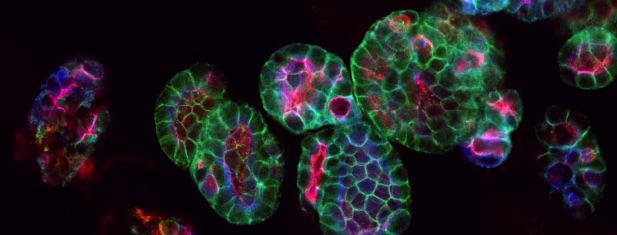
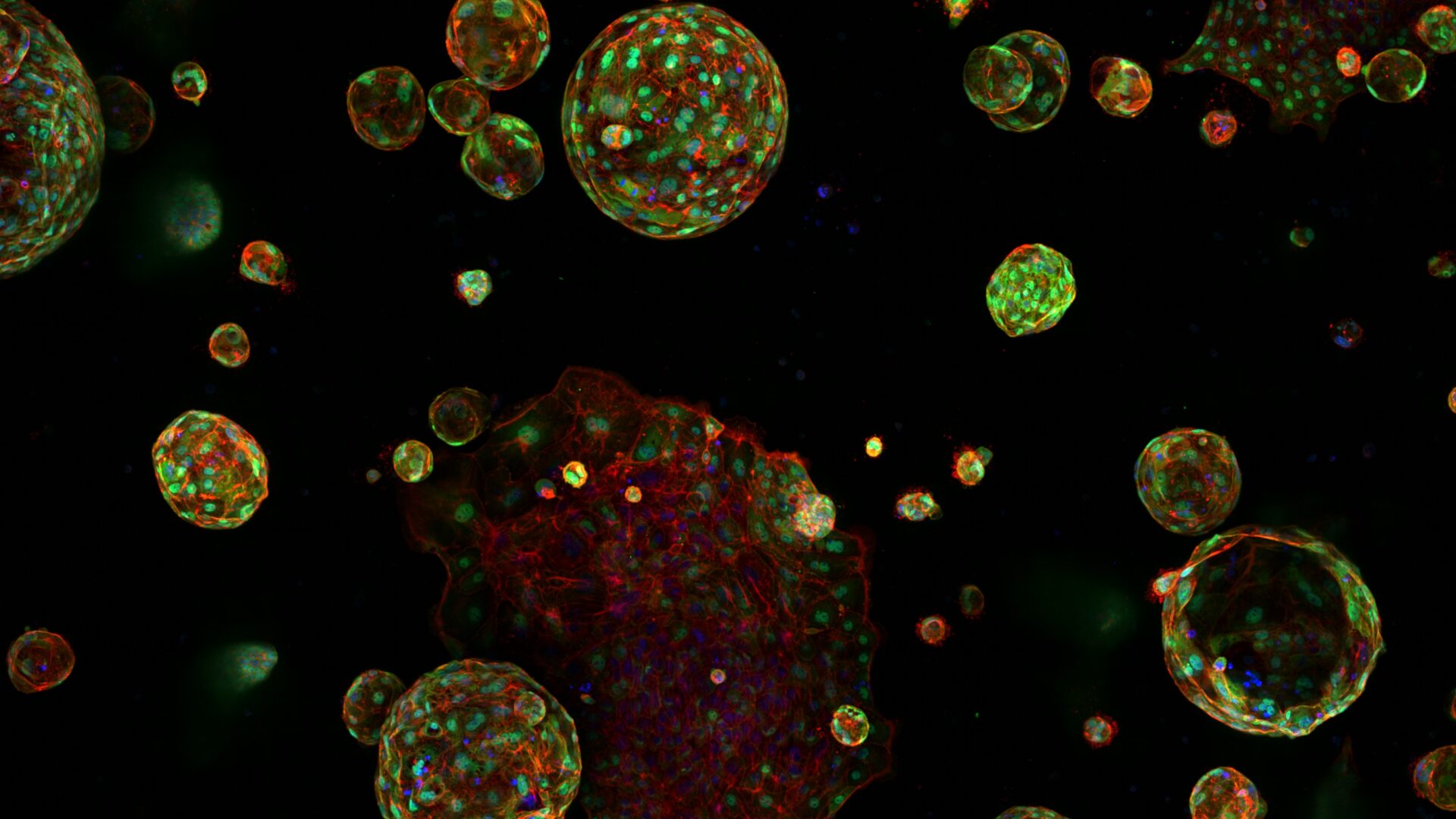
Since 2010 we have been supporting the Institute to utilise High Content Screening to numerate structural biology and response. Our services permit the imaging and analysis of the phenotype of multiple cell populations so as to study cellular responses, for applications including drug discovery and 3D culture (spheroids and organoids).
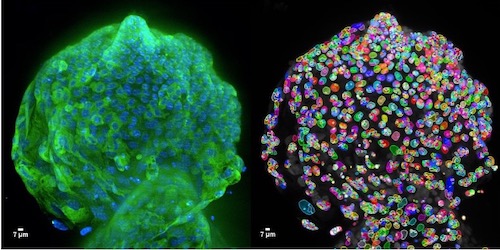
High Content Screening Systems
High throughput and high content screening has emerged as a transformative technology, enabling the comprehensive analysis of cellular and molecular characterisation. By integrating automated microscopy, sophisticated image analysis, and advanced data interpretation, the process provides a powerful platform for examining the intricate mechanisms driving cancer biology by imaging all samples and using segmentation and classifier algorithms. HTS/HCS is employed to uncover insights into tumour heterogeneity, signalling pathways, drug resistance, and the identification of therapeutic targets. This approach allows researchers to conduct phenotypic screening with unparalleled accuracy. Utilising fluorescent markers, live-cell imaging, and multiplexing capabilities, HTC/HCS facilitates the simultaneous analysis of multiple parameters, such as cell morphology, protein localization, and molecular interactions.
PerkinElmer Opera Phenix
A spinning disk based confocal screening system with a four dedicated sCMOS cameras, one each per detection channel. Objectives lenses from 1.25-x60 (water immersion). Automation allows loading of multi-well plates at room temperature or from an incubator. Designed for 3D/4D phenotypic screening
Other systems include:
- PerkinElmer Operetta CLS, a fluorescence wide-field based illumination system for screening system multi-well plate
- PerkinElmer Harmony 3D, for the analysis of organoids and spheroids
- Napari, CellProfiler and other open-source software for 2D/3D analysis
The facility is not isolated in its outlook; it values working alongside other facilities and researchers in developing novel workflows.
Steve Bagley
Team Leader, VIA Core Facility
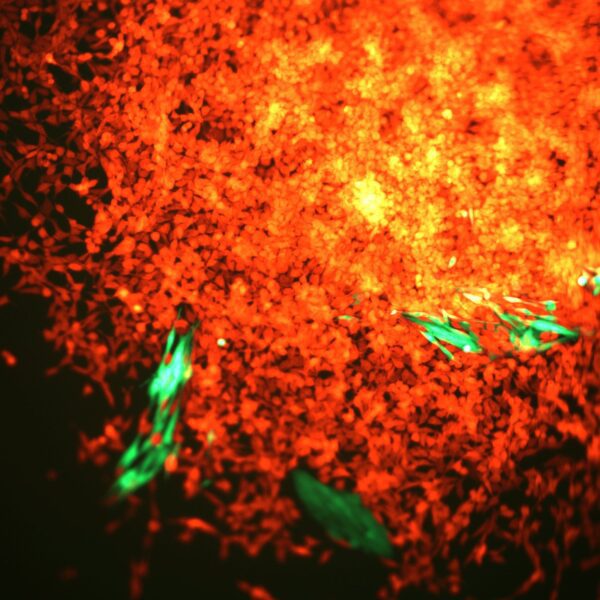
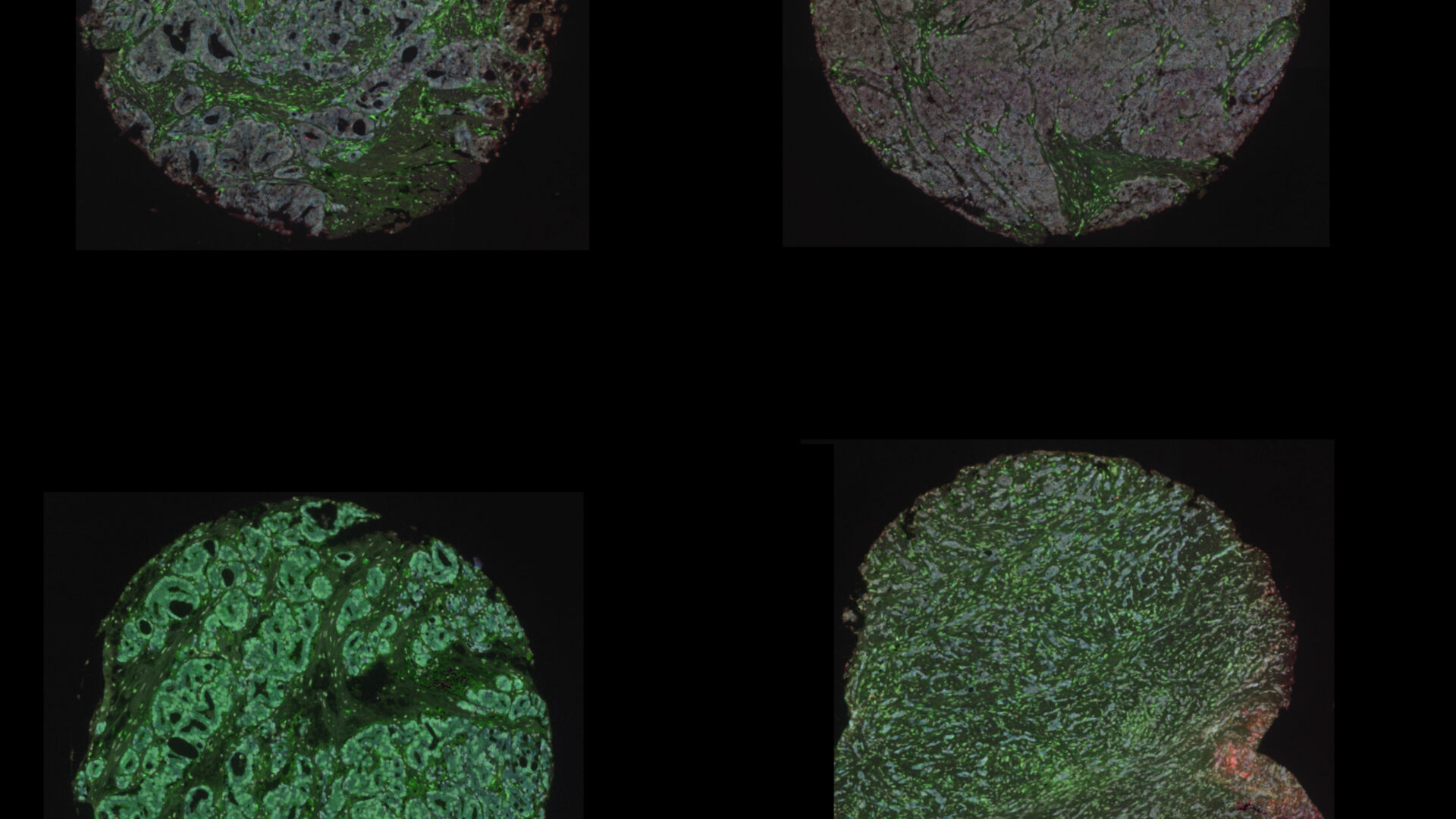
Digital Pathology and Analysis
Since 2008 the institute has been at the cutting edge of whole slide histology and works extremely closely with the Histology Facility. Automated digitisation of whole histology slides for the visualisation of multiplexed tissue (bright-field and fluorescence) at x400 total magnification.
- Olympus VS120, brightfield, polarised and fluorescence (5 colour) on standard slides, able to batch up to 150 slides
- Olympus VS200, brightfield, phase, polarised and fluorescence (5 signal and 7 signal Kromnigon spectra-split) on standard and mega-slides, able to batch up to 200 slides
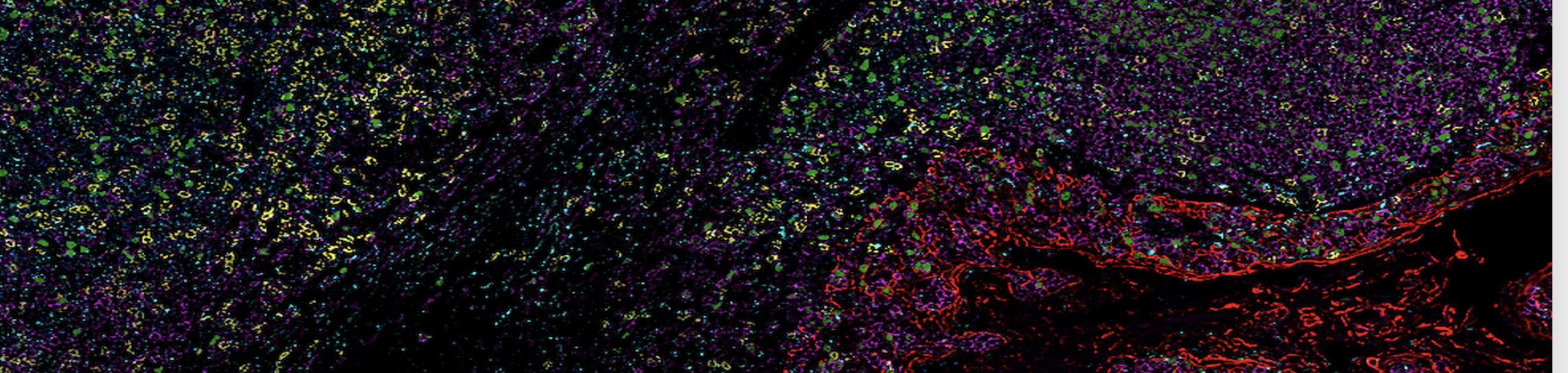
- Akoya Phenocycler Fusion II, to achieve high dimensionality by visualising up to 50 fluorescent labels on a 22x22mm coverslip
- HALO server, analysis software to permit single cell phenotypic analysis
- QuPath, cross-platform, open source software for digital pathology and whole slide image analysis
Frequently Asked Questions
The team are located on the 4th floor of the Paterson building, Steve’s office is 54-04-33
Absolutely, always best to discuss your project with the facilities before you begin. We can discuss panel design, fluorophore choice, the most appropriate workflow or instrument, which analysis tool is suitable. We also suggest you talk to the other facilities, and a bioinformatician.
We will supply links to documents so that you have some knowledge of the theory of the instruments.
Full practical training is given within the facility.
Yes … however we ask you to be present if specific fields of view are required, for setting imaging parameters (exposure etc).
For tissue-based, whole slide imaging, absolutely.
Very much so, we can assist you in developing a new workflow, or we can do some of the groundwork to investigate viability, and provide help in getting a first dataset.
If you are based on the campus, you will require a PPMS account first [https://ppms.eu/man/areq/?pf=3].
All training and instrument requests are handled though the PPMS system
Please contact the facility for full details. Costs cover instrument service contracts, consumables, replacement of some parts and staff time.
Once you are trained and demonstrate competency, you may use the facility 24/7
Scientific Computing securely store all our data and metadata in read-only format on their servers.
We can arrange for data to be moved to an FTP space if data needs to be sent elsewhere (given correct permissions are in place)
The facility can assist in generating the imaging data and taking the data through image processing/analysis. However, the team cannot assist with bioinformatics.
The Computational Biology Support team within the institute may be able to assist, so do get in touch with them.
The laboratory has been awarded gold Laboratory Efficiency Assessment Framework (LEAF) status. LEAF is a standard set by UCL to improve the sustainability and efficiency of laboratories, so to address the climate and ecological emergencies our world faces through sustainable science. [For more details visit:https://www.ucl.ac.uk/sustainable/take-action/staff-action/leaf-laboratory-efficiency-assessment-framework].
The team have adopted these principles in a way that has not impacted workflows but has enabled the facility to work smarter.
If you have a CRUK MI PPMS account, all documentation detailing the instruments are stored at https://ppms.eu/man/login/?pf=3
Get in touch
Our vision for world leading cancer research in the heart of Manchester
We are a leading cancer research institute within The University of Manchester, spanning the whole spectrum of cancer research – from investigating the molecular and cellular basis of cancer, to translational research and the development of therapeutics.
Our collaborations
Bringing together internationally renowned scientists and clinicians
Scientific Advisory Board
Supported by an international Scientific Advisory Board
Careers that have a lasting impact on cancer research and patient care
We are always on the lookout for talented and motivated people to join us. Whether your background is in biological or chemical sciences, mathematics or finance, computer science or logistics, use the links below to see roles across the Institute in our core facilities, operations teams, research groups, and studentships within our exceptional graduate programme.









A note from the Team Leader – Steve Bagley
Our remit is to support researchers with imaging and irradiation instrumentation, provide quality control and support for image processing/analysis. We also work closely with research teams to develop new techniques and instrumentation.