The Computational Biology Support team (CBS) provides bioinformatics support and collaborative data analysis solutions to researchers across the CRUK Manchester Institute. Our core activities reside at the interface between biology, computer science and statistics, particularly in the application of high-throughput omics technologies. The primary goal of the CBS team is to build and maintain an infrastructure that enables the application of advanced bioinformatics analysis to cutting-edge scientific research within CRUK MI.
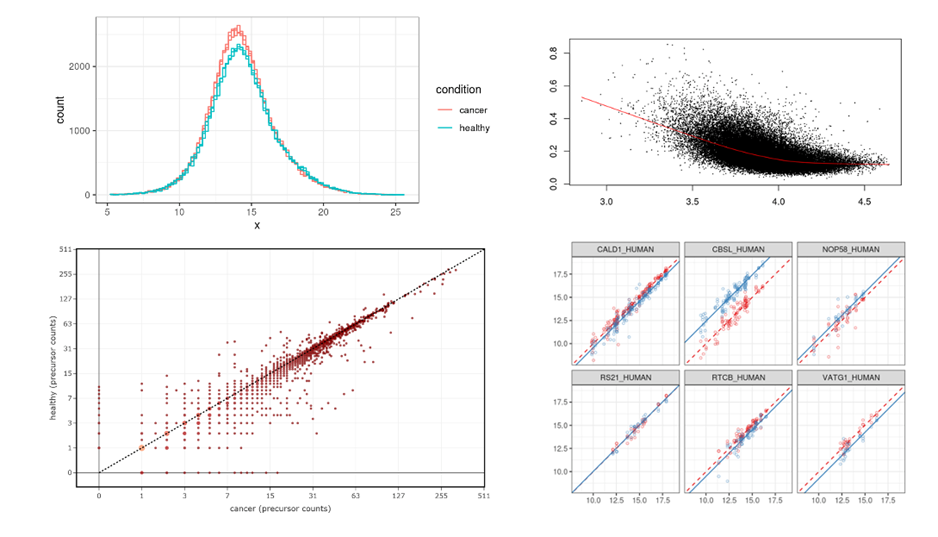
A major aspect of our work has been to develop pipelines and computational approaches for analysing high-throughput datasets generated from next generation sequencing (NGS) and mass-spectrometry platforms. Most of the workflows that we developed utilise a combination of open-source and custom-built software tools running on the on-site high performance computing facility provided by the Scientific Computing team. Working closely with the SciCom team, we are currently building automated pipelines to handle routine tasks associated with the pre-processing of deep sequencing dataset, freeing valuable time for the more challenging downstream analysis and results interpretation.
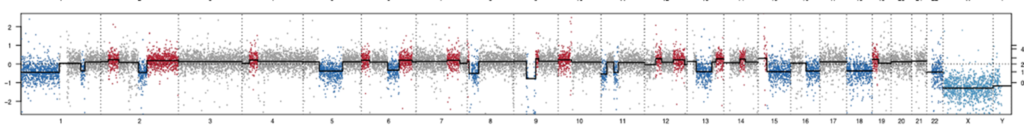
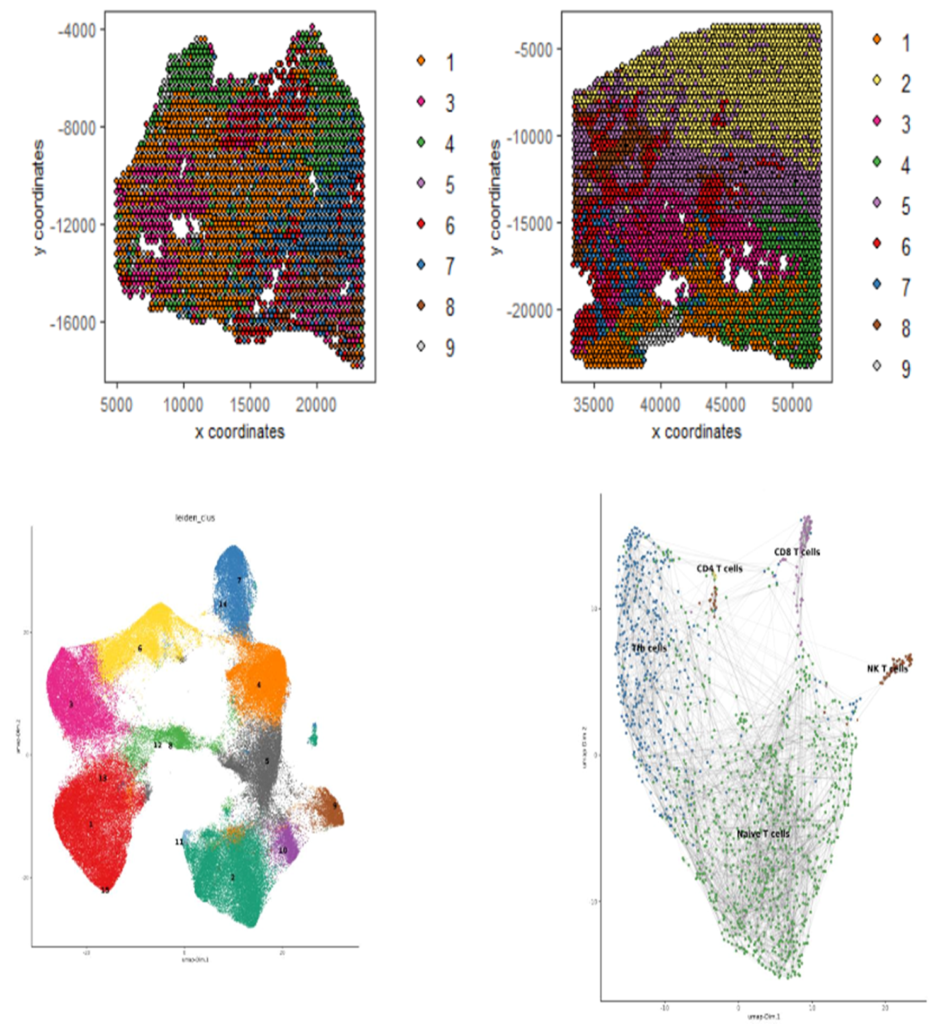
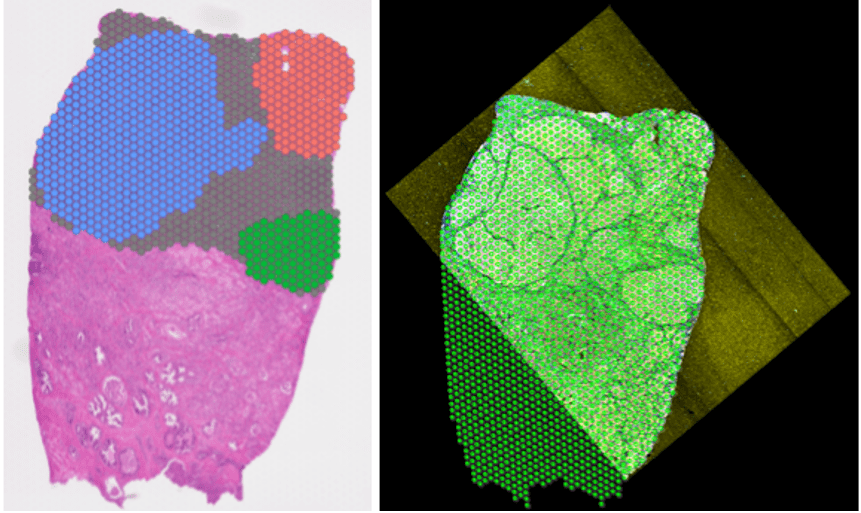
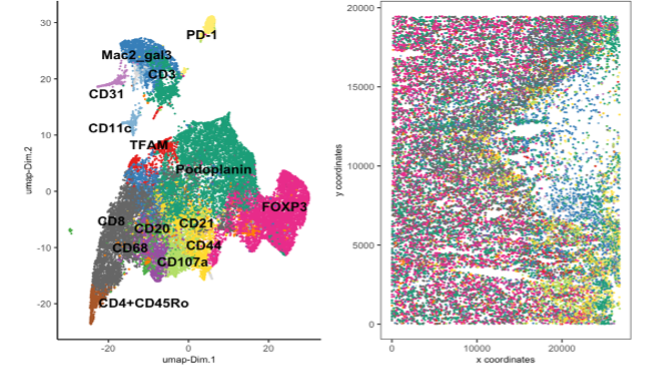
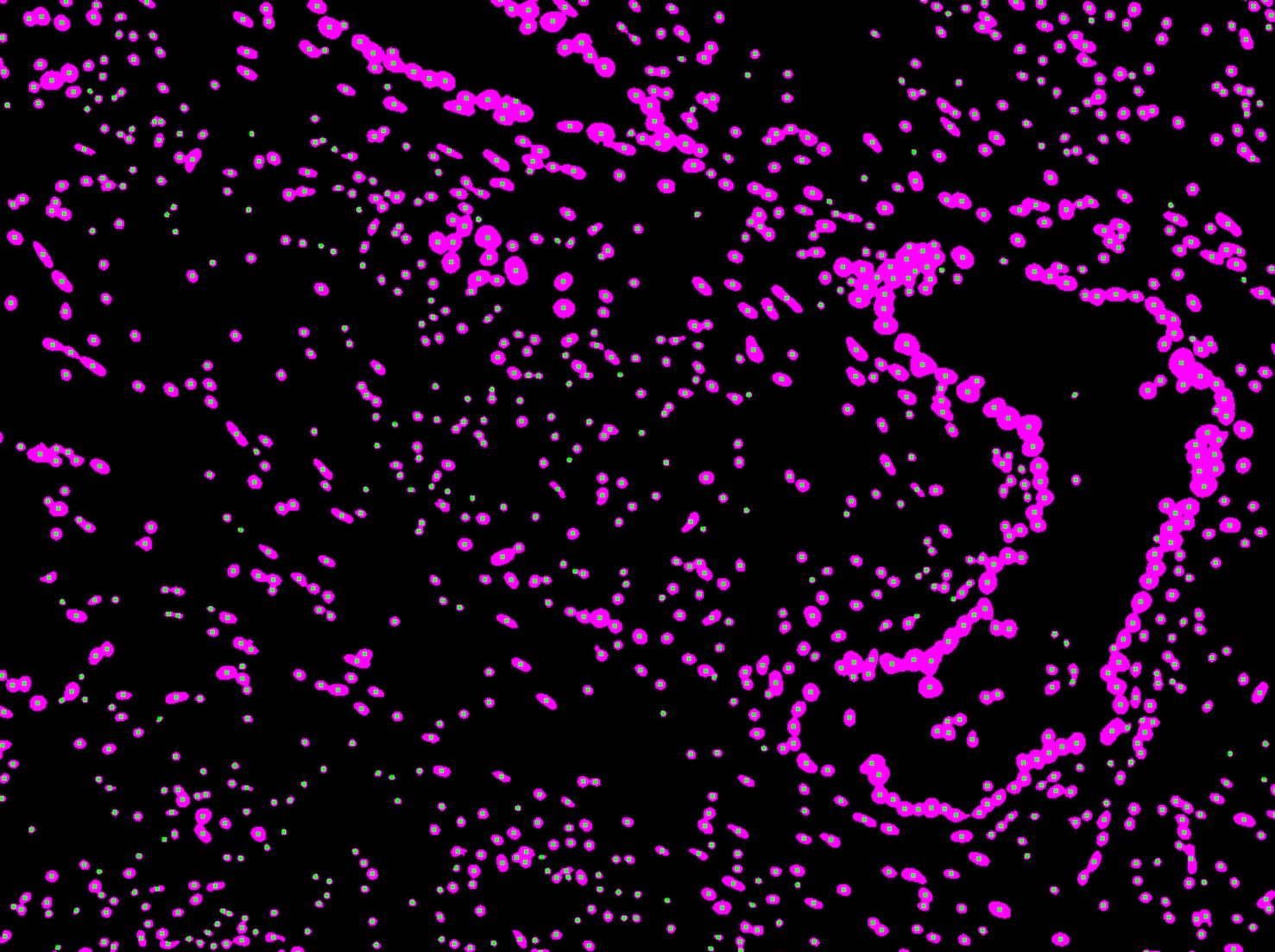
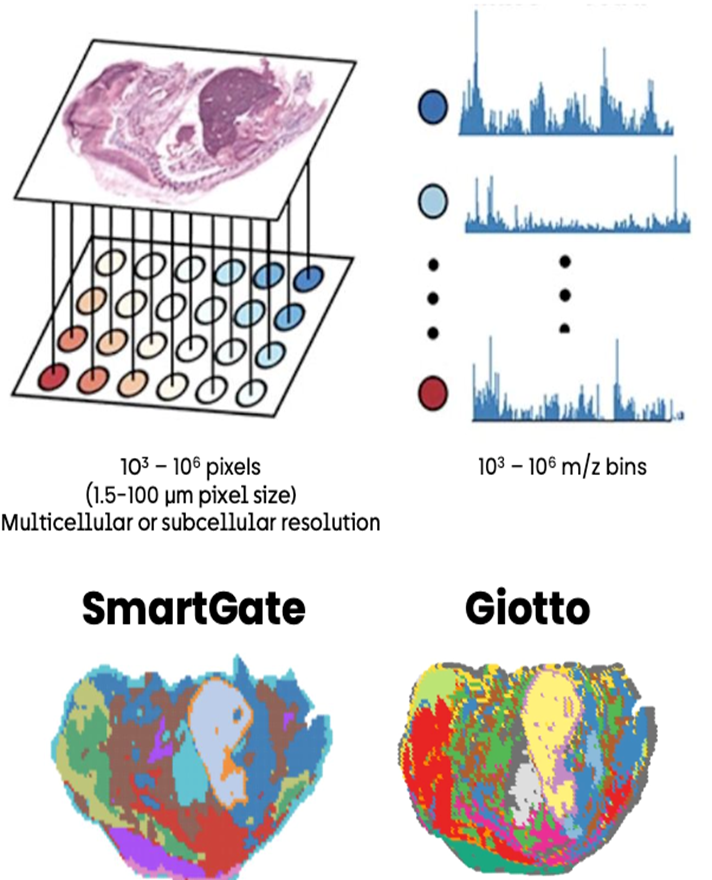
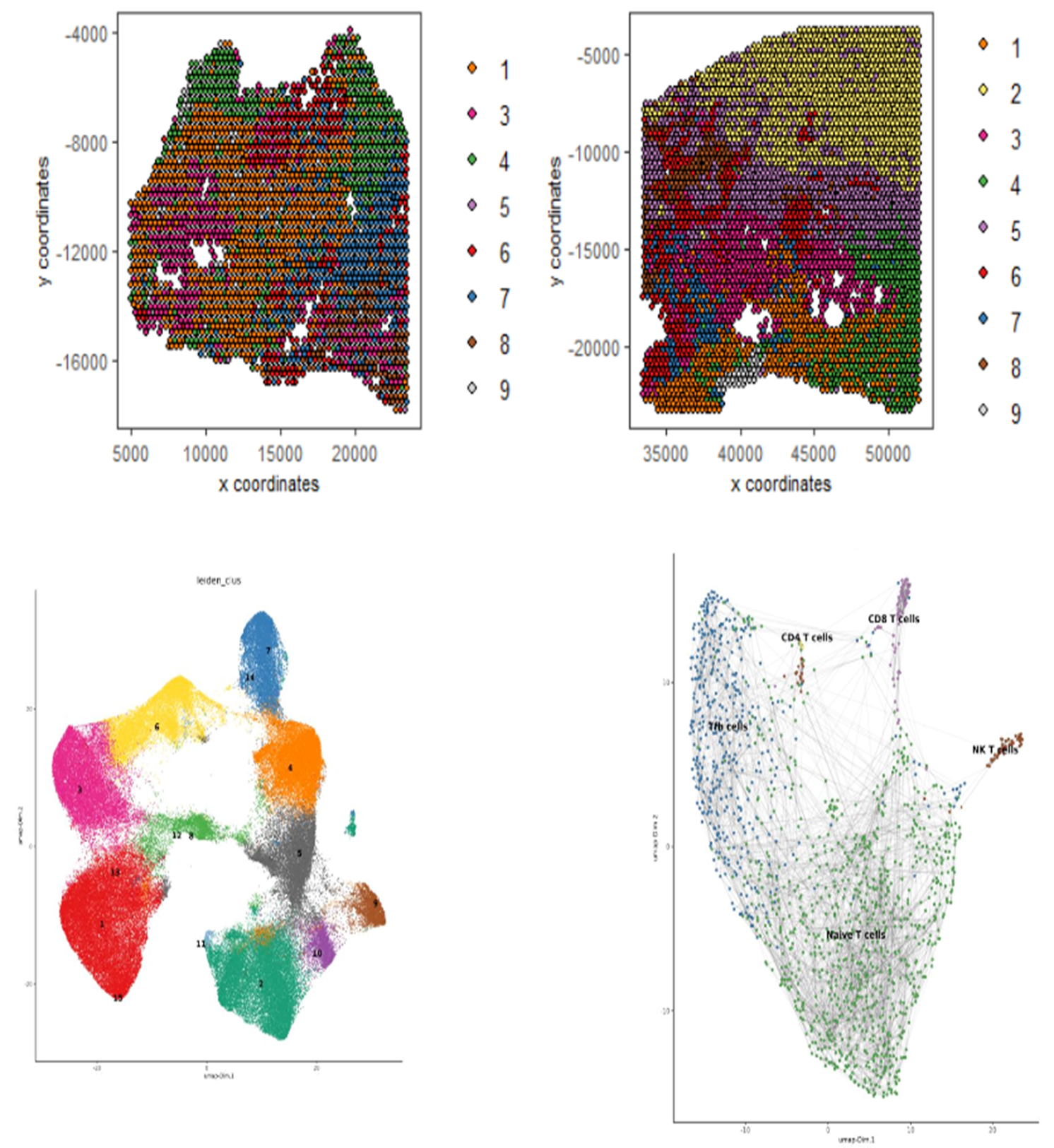
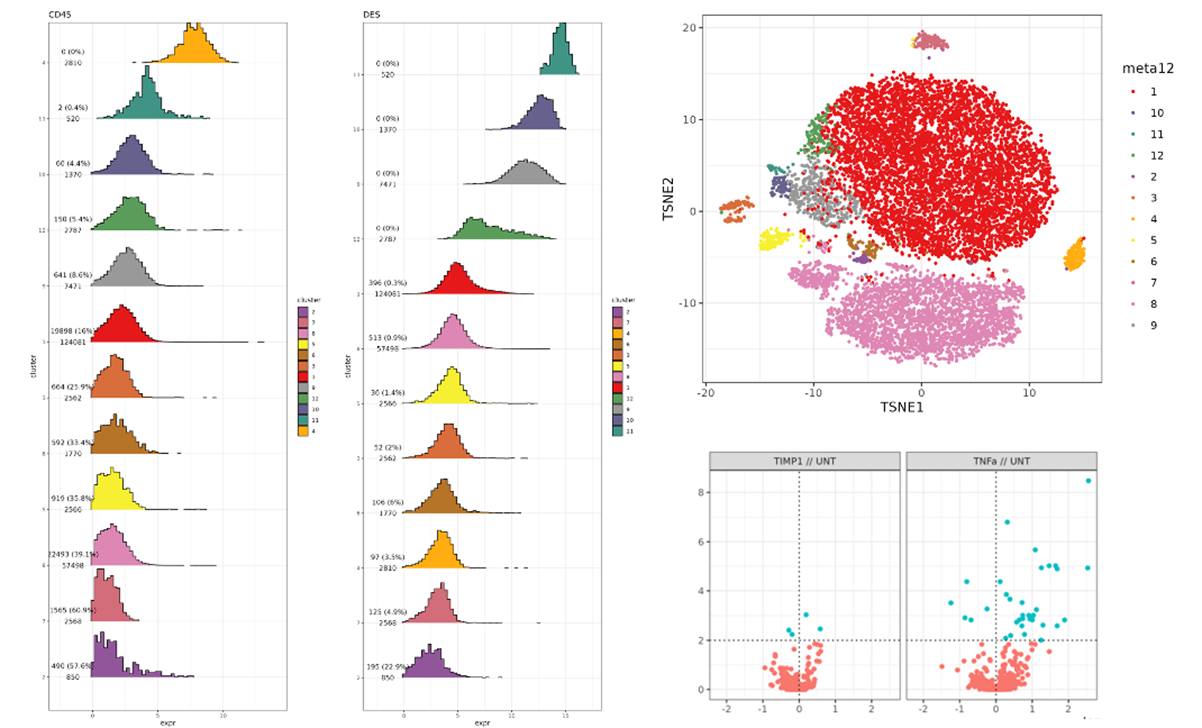
Currently, the CBS team provide computational biology and bioinformatics support to the research groups at the Institute on genomics, epigenomics, transcriptomics, and highly multiplexed image analysis, including spatially resolved transcriptomic and proteomic data analysis. We utilise state-of-the-art computational tools, including machine learning and AI driven deep learning tools for spatially resolved cellular and subcellular multi-omics and multi-modal data analysis and integration.
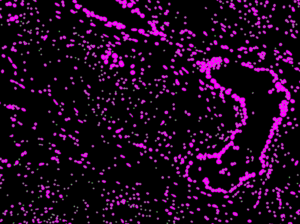
They offer guidance on experimental design, conduct data analysis, and provide bioinformatics training across the Institute in various areas including:
- Genomics, Transcriptomics, Proteomics, Metabolomics and Highly-multiplexed Imaging analysis
- Bulk and Single-Cell Genomics and Transcriptomics/Multi-modal Analysis
- Nanopore direct RNA sequencing analysis
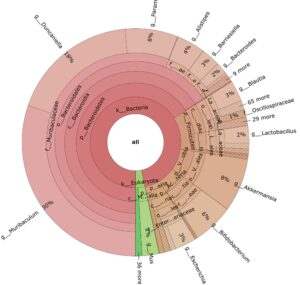
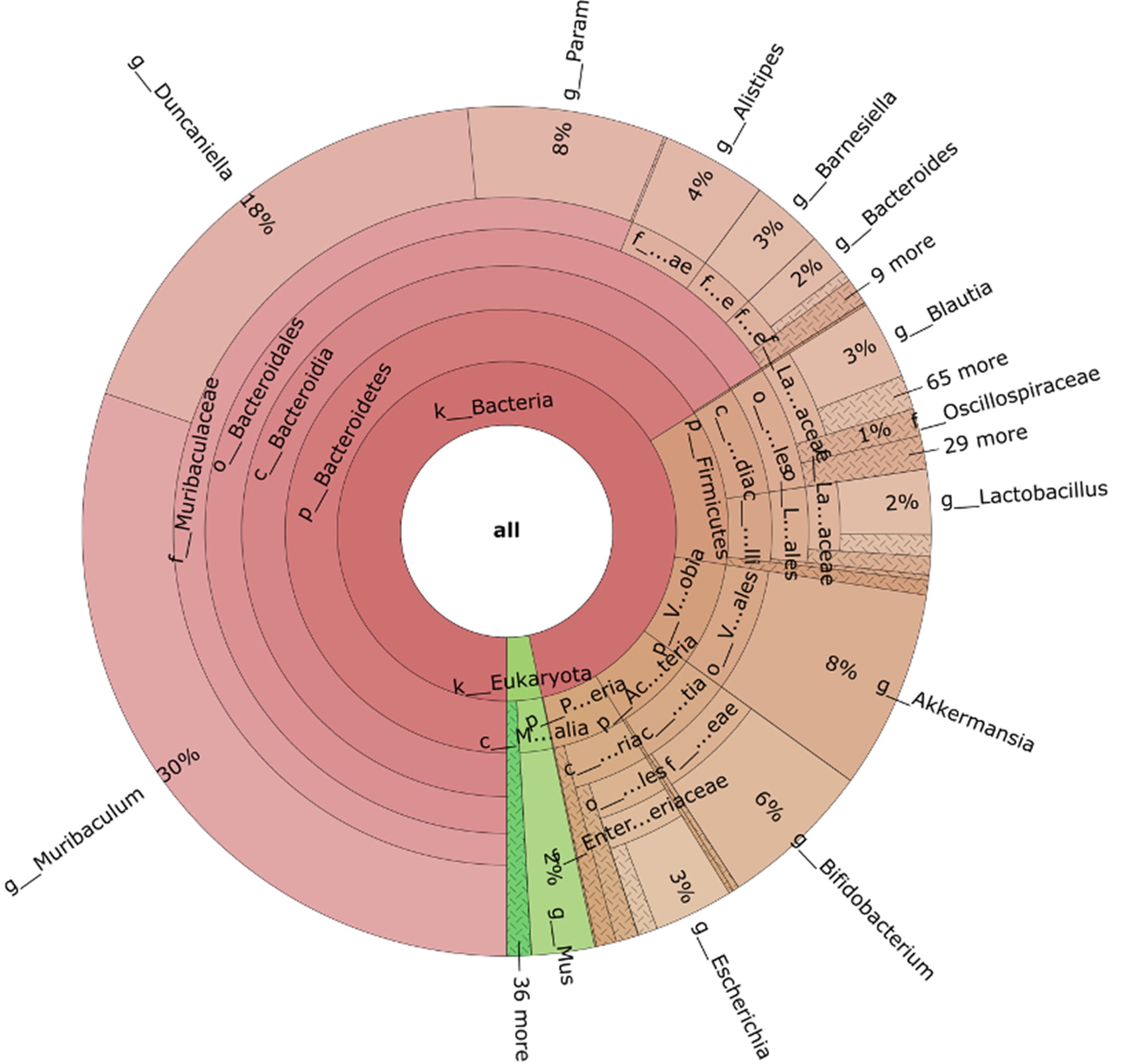
- Metagenomics
- Image processing and segmentation
CBS also provide support for collecting data from publicly available data sets, such as TCGA/dbGAP and from published articles, working with research teams to plug this content into new workflows.
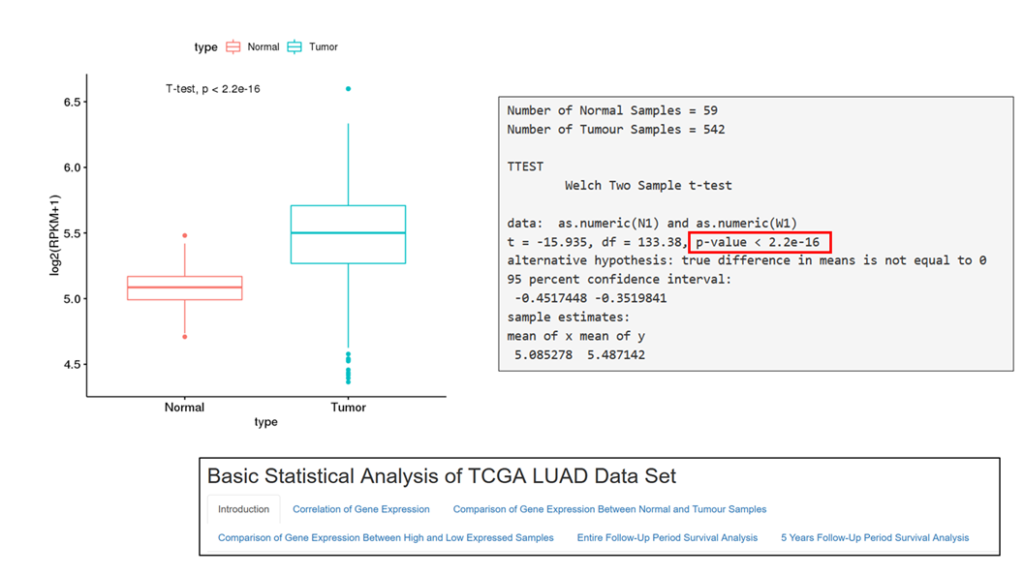
The CBS team support data management, processing and analysis from a range of spatial transcriptomic and proteomic platforms, including workflows for:
- 10X Visium/Xenium
- NanoString GeoMx DSP/CosMx MSI
- mIHC
- mIF
- CODEX
Meet the Computational Biology Support team
Core Facility Manager

Computational Biologist
Computational Biologist

Computational Biologist
Get in touch
Our vision for world leading cancer research in the heart of Manchester
We are a leading cancer research institute within The University of Manchester, spanning the whole spectrum of cancer research – from investigating the molecular and cellular basis of cancer, to translational research and the development of therapeutics.
Our collaborations
Bringing together internationally renowned scientists and clinicians
Scientific Advisory Board
Supported by an international Scientific Advisory Board
Careers that have a lasting impact on cancer research and patient care
We are always on the lookout for talented and motivated people to join us. Whether your background is in biological or chemical sciences, mathematics or finance, computer science or logistics, use the links below to see roles across the Institute in our core facilities, operations teams, research groups, and studentships within our exceptional graduate programme.


A note from the Team Leader – Sudhakar Sahoo
The CBS team has the broad expertise to analyse and interrogate genomic, epigenomic, transcriptomic and proteomic datasets at bulk, from single cell to spatial resolutions. They produce workflows and pipelines to perform a variety of high throughput omics, multi-omics and multimodal data analysis and integration supporting many of our research groups.