Facilities
Facilities
Scientific Research Services across the Institute
The scientific research services in the Cancer Research UK Manchester Institute are overseen by Stuart Pepper, our Chief Laboratory Officer. Each Core Facility is led by a dedicated team leader who is always available and willing to discuss the services they provide, including the implementation of new techniques and approaches tailored to specific research projects and investigations.
By working closely with Institute scientists, the Core Facilities are able to continually develop their services, offering support for the evolving research interests of the Institute. The Facilities provide a diverse range of technologies that are selected to support the Institute research programs.
Each Facility Manager is supported by a team of expert staff who can provide support and training to our early career scientists.
The ethos of the core facilities is summed up by collaboration. This applies both to working collaboratively with research groups to develop new technologies and to the Core Facilities working collaboratively together to develop workflows that span technologies.
Our Vision and Values
Service Development Through Collaboration
The core facilities span the traditional areas of Molecular Biology, Histology, Imaging, Mass Spectrometry and in vivo technologies, however new services are frequently delivered as a result of collaboration between facilities to provide seamless workflows that span across these traditionally separate disciplines.
This applies particularly to the Imaging and bioinformatic capabilities: the imaging team work closely with histology and molecular biology teams on the development of spatial workflows as described below whilst the bioinformatics team work closely with most core facilities, but particularly the Scientific Computing facility to ensure that data analysis workflows are developed to run efficiently on our HPC platform.
New platforms are also developed in close collaboration with research groups. Initial work on the implementation of the Helios platform for mass cytometry was a collaboration between the Systems Oncology group and the flow cytometry team. The combination of a research scientist working with the core facility team allowed rapid progress to demonstrate the power of the technology and develop the platform as a service offering. Similar interactions happen with our BRU experimental team who work closely with researchers who can bring expertise in novel procedures, allowing the BRU teams then to develop in house expertise that can be shared across research groups.

Broader, Faster, Deeper
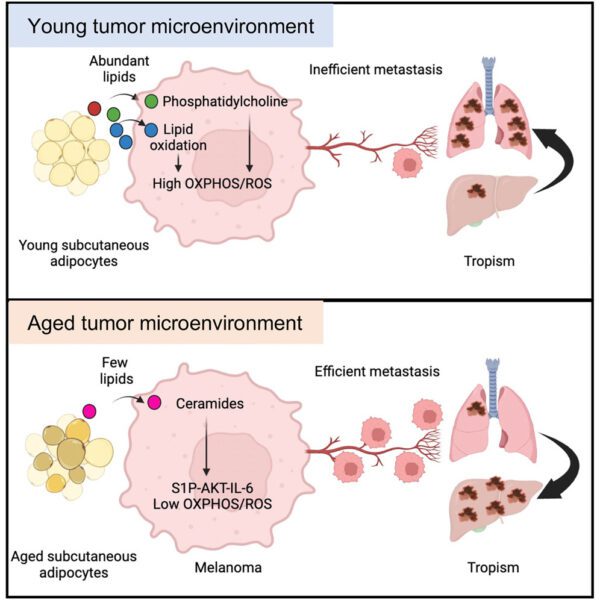
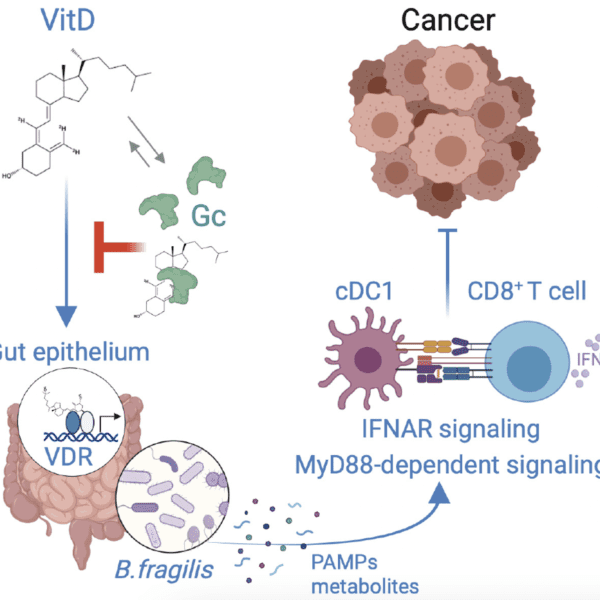
As the sheer complexity of cancer biology has become clearer over the last decade, there has been a continual drive to be able to look in more detail at biological systems; this means wanting to quantitate more proteins, study large numbers of individual cells and look at the spatial relationships between tumours and normal tissue.
With the colocation of our histology, imaging, genomics and data management and analysis capabilities we have been well positioned to implement a number of workflows in the rapidly evolving field of spatial -omics that support this requirement for high resolution analysis of biological samples. Currently the facilities support 10X Genomics Visium, Nanostring GeoMx and STOmics Stereoseq. Further high multiplex platforms for Histology are also supported, such as the Phenocycler and Ultivue systems.

Sustainability and Core Facilities
Research is an energy and resource intensive activity, consequently there is intense interest in how we can improve the sustainability of research activity. Core facilities make an important contribution to sustainability in several ways: by ensuring equipment is well maintained we get the optimum lifespan from each item; by making it accessible to multiple groups we ensure a high level of usage of equipment, and by offering expertise we can help to ensure that each experiment is a success, reducing the risk of wastage if experiments fail.
Alongside the research groups, all the core facilities are committed to following the LEAF system; at the end of 2024 all facilities were at a minimum of Bronze and some had already reached Silver and Gold level. The aim is to have all facilities at Gold level before the end of 2025.
Across the Institute a number of projects are being explored around how to reduce waste from single use plastics either by looking at alternatives, or opting for recycling. You can find out more about the ways in which we are incorporating sustainable thinking into our daily work on our
Sustainability page.

Our Core Facilities
By working closely with Institute scientists, the core facilities are able to continually develop their services, offering support for the evolving research interests of the Institute. Alongside end-to-end sample processing, the core facilities also provide training so that our early career scientists gain valuable expertise.

Biological Mass Spectrometry
Protein and peptide mass spectrometry analysis

BRU Experimental Team
Experimental animal models supporting cancer research
Why choose Cancer Research UK Manchester Institute?
The Cancer Research UK Manchester Institute, an Institute of The University of Manchester, is a world-leading centre for excellence in cancer research. The Institute is core funded by Cancer Research UK (www.cancerresearchuk.org),
the largest independent cancer research organisation in the world.
We are partnered with The Christie NHS Foundation Trust, one of the largest cancer treatment centres in Europe, which is located adjacent to the CRUK MI Manchester Institute in South Manchester. These factors combine to provide an exceptional environment in which to pursue basic, translational and clinical research programmes.