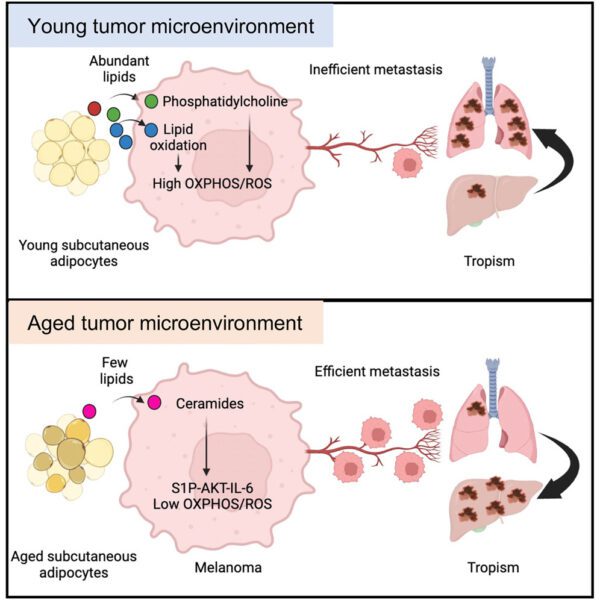
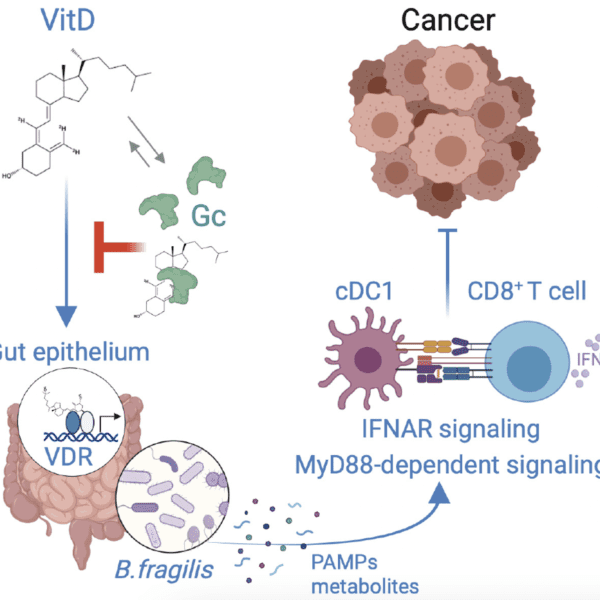
Article highlights & insights
Former Institute research group members previously identified that mutant p53 expression in cancer cells promotes engulfment of neighbouring cancer cells to form cell-in-cell (CIC) structures. This process gave mutant p53 cells an advantage in tumour formation in mouse xenograft experiments. TP53 can be found mutated at nearly every amino acid in cancers and mutant p53 expression is associated with various GOF (Gain-of-function) processes, including cancer cell invasion, metastasis, stemness and drug resistance.
In this publication, they identified SH3BGRL (Src homology 3 binding glutamate rich protein like) as a mutant p53-regulated gene and investigated to what extent SH3BGRL expression and cell engulfment are responsible for mutant p53-dependent anchorage-independent growth and chemoresistance.
Here they demonstrate that mutant p53 expression drives cell engulfment in which the mutant p53 host cell moves in the direction of the target internal cell to form CIC structures. This is therefore more reminiscent of cell engulfment rather than cell entosis, in which cells invade into host cells.
Using NGS (Next Generation Sequencing), they identified novel target genes of mutant p53 and demonstrate that cell engulfment requires SH3BGRL expression. They generated mutant p53 and p53 KO cell lines that stably overexpressed SH3BGRL and determined that SH3BGRL promotes etoposide resistance in mutant p53 cells and anchorage-independent growth independent of mutant p53 expression.
Through FACS sorting of pure cell engulfing (CIC) populations, we could also show that engulfing cells have an enhanced etoposide resistance. These data suggest that SH3BGRL and cell engulfment are required for certain GOFs of mutant p53.
Former Institute research group members previously identified that mutant p53 expression in cancer cells promotes engulfment of neighbouring cancer cells to form cell-in-cell (CIC) structures. This process gave mutant p53 cells an advantage in tumour formation in mouse xenograft experiments. TP53 can be found mutated at nearly every amino acid in cancers and mutant p53 expression is associated with various GOF (Gain-of-function) processes, including cancer cell invasion, metastasis, stemness and drug resistance.
In this publication, they identified SH3BGRL (Src homology 3 binding glutamate rich protein like) as a mutant p53-regulated gene and investigated to what extent SH3BGRL expression and cell engulfment are responsible for mutant p53-dependent anchorage-independent growth and chemoresistance.
Here they demonstrate that mutant p53 expression drives cell engulfment in which the mutant p53 host cell moves in the direction of the target internal cell to form CIC structures. This is therefore more reminiscent of cell engulfment rather than cell entosis, in which cells invade into host cells.
Using NGS (Next Generation Sequencing), they identified novel target genes of mutant p53 and demonstrate that cell engulfment requires SH3BGRL expression. They generated mutant p53 and p53 KO cell lines that stably overexpressed SH3BGRL and determined that SH3BGRL promotes etoposide resistance in mutant p53 cells and anchorage-independent growth independent of mutant p53 expression.
Through FACS sorting of pure cell engulfing (CIC) populations, we could also show that engulfing cells have an enhanced etoposide resistance. These data suggest that SH3BGRL and cell engulfment are required for certain GOFs of mutant p53.