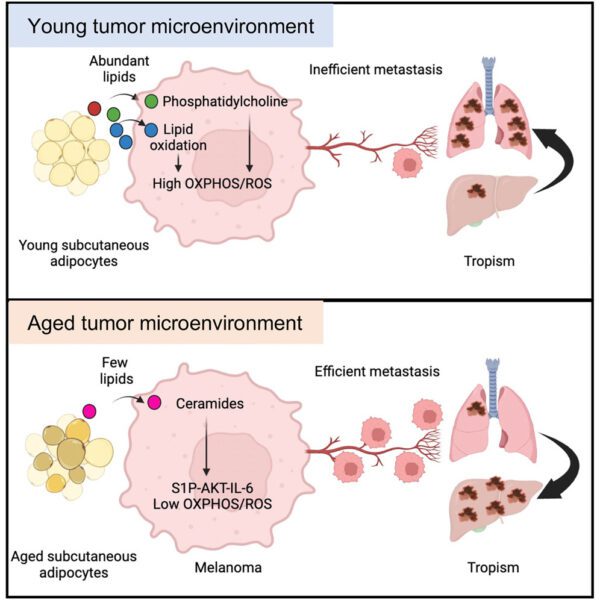
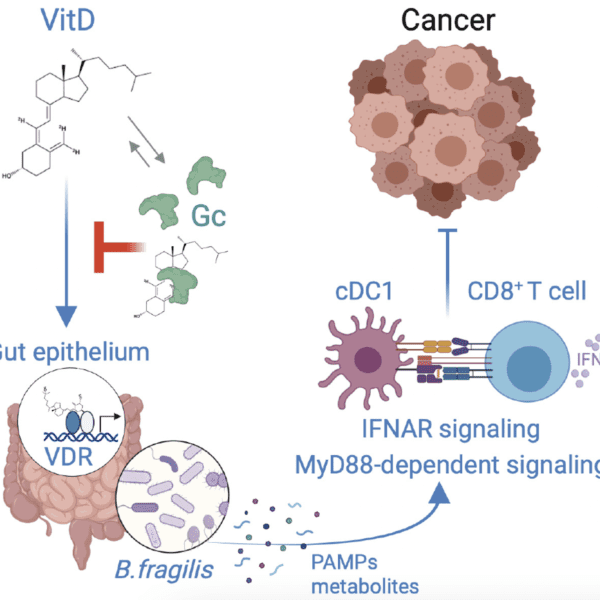
Article highlights & insights
While the key aspects of genetic evolution and their clinical implications in clear cell renal-cell carcinoma (ccRCC) are well-documented, how genetic features co-evolve with the phenotype and tumour microenvironment (TME) remains elusive. Here, through joint genomic-transcriptomic analysis of 243 samples from 79 patients recruited to the TRACERx Renal study, the group identify pervasive non-genetic intratumour heterogeneity, with over 40% not attributable to genetic alterations. By integrating tumour transcriptomes and phylogenetic structures, they observe convergent evolution to specific phenotypic traits, including cell proliferation, metabolic reprogramming and overexpression of putative cGAS-STING repressors amid high aneuploidy. The team also uncovered a co-evolution between the tumour and the T cell repertoire, as well as a longitudinal shift in the TME from an anti-tumour to an immunosuppressive state, linked to the acquisition of recurrently late ccRCC drivers 9p loss and SETD2 mutations. This study reveals clinically-relevant and hitherto underappreciated non-genetic evolution patterns in ccRCC.
While the key aspects of genetic evolution and their clinical implications in clear cell renal-cell carcinoma (ccRCC) are well-documented, how genetic features co-evolve with the phenotype and tumour microenvironment (TME) remains elusive. Here, through joint genomic-transcriptomic analysis of 243 samples from 79 patients recruited to the TRACERx Renal study, the group identify pervasive non-genetic intratumour heterogeneity, with over 40% not attributable to genetic alterations. By integrating tumour transcriptomes and phylogenetic structures, they observe convergent evolution to specific phenotypic traits, including cell proliferation, metabolic reprogramming and overexpression of putative cGAS-STING repressors amid high aneuploidy. The team also uncovered a co-evolution between the tumour and the T cell repertoire, as well as a longitudinal shift in the TME from an anti-tumour to an immunosuppressive state, linked to the acquisition of recurrently late ccRCC drivers 9p loss and SETD2 mutations. This study reveals clinically-relevant and hitherto underappreciated non-genetic evolution patterns in ccRCC.
Institute Authors
Groups
Group leader
Research topics & keywords
Our Research
Our research spans the whole spectrum of cancer research from cell biology through to translational and clinical studies
Research Groups
Our research groups study many fundamental questions of cancer biology and treatment
Our Facilities
The Institute has outstanding core facilities that offer cutting edge instruments and tailored services from expert staff
Latest News & Updates
Find out all our latest news