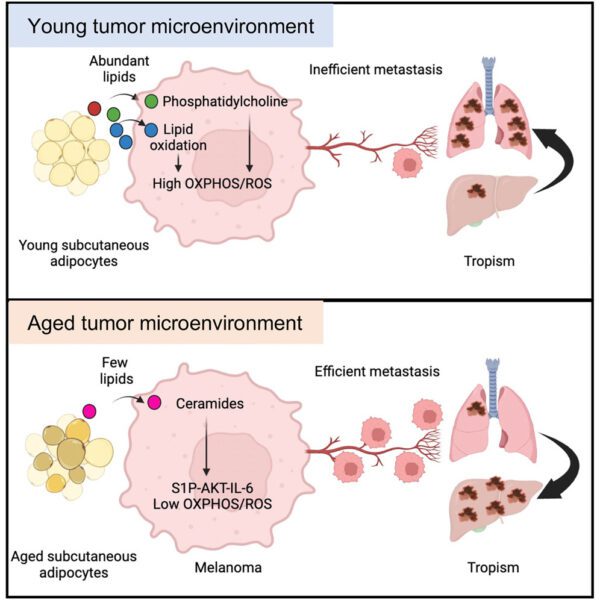
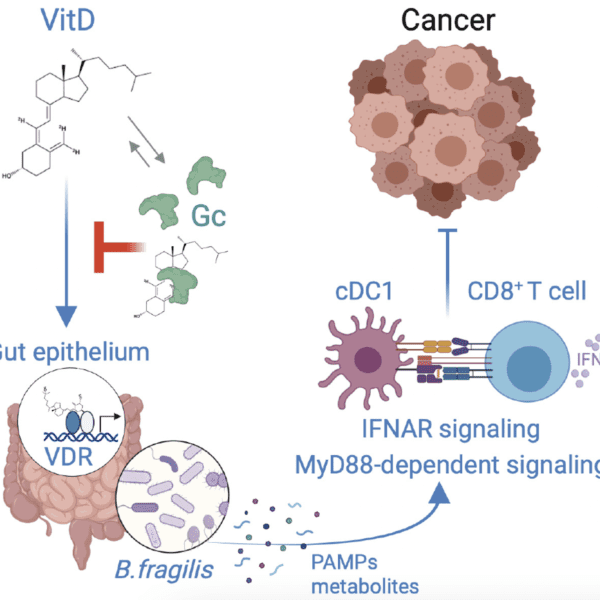
Article highlights
As we age, our tissues are repeatedly challenged by mutational insult, yet cancer occurrence is a relatively rare event. Cells carrying cancer-causing genetic mutations compete with normal neighbors for space and survival in tissues. However, the mechanisms underlying mutant-normal competition in adult tissues and the relevance of this process to cancer remain incompletely understood. Here, we investigate how the adult pancreas maintains tissue health in vivo following sporadic expression of oncogenic Kras (KrasG12D), the key driver mutation in human pancreatic cancer. We find that when present in tissues in low numbers, KrasG12D mutant cells are outcompeted and cleared from exocrine and endocrine compartments in vivo. Using quantitative 3D tissue imaging, we show that before being cleared, KrasG12D cells lose cell volume, pack into round clusters, and E-cadherin-based cell-cell adhesions decrease at boundaries with normal neighbors.
We identify EphA2 receptor as an essential signal in the clearance of KrasG12D cells from exocrine and endocrine tissues in vivo. In the absence of functional EphA2, KrasG12D cells do not alter cell volume or shape, E-cadherin-based cell-cell adhesions increase and KrasG12D cells are retained in tissues. The retention of KRasG12D cells leads to the early appearance of premalignant pancreatic intraepithelial neoplasia (PanINs) in tissues. Our data show that adult pancreas tissues remodel to clear KrasG12D cells and maintain tissue health. This study provides evidence to support a conserved functional role of EphA2 in Ras-driven cell competition in epithelial tissues and suggests that EphA2 is a novel tumor suppressor in pancreatic cancer.
As we age, our tissues are repeatedly challenged by mutational insult, yet cancer occurrence is a relatively rare event. Cells carrying cancer-causing genetic mutations compete with normal neighbors for space and survival in tissues. However, the mechanisms underlying mutant-normal competition in adult tissues and the relevance of this process to cancer remain incompletely understood. Here, we investigate how the adult pancreas maintains tissue health in vivo following sporadic expression of oncogenic Kras (KrasG12D), the key driver mutation in human pancreatic cancer. We find that when present in tissues in low numbers, KrasG12D mutant cells are outcompeted and cleared from exocrine and endocrine compartments in vivo. Using quantitative 3D tissue imaging, we show that before being cleared, KrasG12D cells lose cell volume, pack into round clusters, and E-cadherin-based cell-cell adhesions decrease at boundaries with normal neighbors.
We identify EphA2 receptor as an essential signal in the clearance of KrasG12D cells from exocrine and endocrine tissues in vivo. In the absence of functional EphA2, KrasG12D cells do not alter cell volume or shape, E-cadherin-based cell-cell adhesions increase and KrasG12D cells are retained in tissues. The retention of KRasG12D cells leads to the early appearance of premalignant pancreatic intraepithelial neoplasia (PanINs) in tissues. Our data show that adult pancreas tissues remodel to clear KrasG12D cells and maintain tissue health. This study provides evidence to support a conserved functional role of EphA2 in Ras-driven cell competition in epithelial tissues and suggests that EphA2 is a novel tumor suppressor in pancreatic cancer.
Groups
Group leader
Research topics & keywords
Meet the Research Team
The Cancer Origins group is dedicated to understanding how cancer begins, specifically, how oncogenic cells overcome normal tissue restraints, gain malignant potential, and initiate tumour formation. We are interested in how environmental exposures, such as air pollutants, shape the tissue microenvironment to support the expansion of latent oncogenic cells and promote lung cancer

Institute Fellow
All publications
https://doi.org/10.1038/s44161-025-00740-z
Single-cell profiling reveals three endothelial-to-hematopoietic transitions with divergent isoform expression landscapes
11 November 2025
Institute Authors (6)
Robert Sellers, John Weightman, Wolfgang Breitwieser, Natalia Moncaut, Michael Lie-a-ling, Georges Lacaud
Labs & Facilities
Computational Biology Support, Molecular Biology, Genome Editing and Mouse Models
Research Group
Stem Cell Biology
11 November 2025
https://doi.org/10.1136/jitc-2025-012527
Systemic immunosuppression from ultraviolet radiation exposure inhibits cancer immunotherapy
31 October 2025
Institute Authors (4)
Isabella Mataloni, Antonia Banyard, Garry Ashton, Amaya Virós
Labs & Facilities
Mass and Flow Cytometry, Histology
Research Group
Skin Cancer & Ageing
31 October 2025
https://aacrjournals.org/cancerdiscovery/article/doi/10.1158/2159-8290.CD-24-1224/766638/Glucocorticoids-Unleash-Immune-dependent-Melanoma
Glucocorticoids Unleash Immune-dependent Melanoma Control through Inhibition of the GARP/TGF β Axis
15 October 2025
Institute Authors (12)
Charles Earnshaw, Poppy Dunn, Shih-Chieh Chiang, Maria Koufaki, Massimo Russo, Kimberley Hockenhull, Erin Richardson, Anna Pidoux, Alex Baker, Richard Reeves, Robert Sellers, Sudhakar Sahoo
Labs & Facilities
Computational Biology Support, Visualisation, Irradiation and Analysis
Research Group
Cancer Inflammation and Immunity
15 October 2025
/wp-content/uploads/2025/09/Annual_Report_2024.pdf
2024 Annual Report
23 September 2025
23 September 2025
https://doi.org/10.1182/blood.2024028033
An in vivo barcoded CRISPR-Cas9 screen identifies Ncoa4-mediated ferritinophagy as a dependence in Tet2-deficient hematopoiesis
4 September 2025
Institute Authors (1)
Justin Loke
Research Group
Myeloid Cancer Biology
4 September 2025
https://doi.org/10.1038/s41467-024-49692-1
Whole genome sequencing refines stratification and therapy of patients with clear cell renal cell carcinoma
15 July 2025
Institute Authors (1)
Samra Turajlić
Research Group
Cancer Dynamics
15 July 2025
Our Research
Our research spans the whole spectrum of cancer research from cell biology through to translational and clinical studies
Research Groups
Our research groups study many fundamental questions of cancer biology and treatment
Our Facilities
The Institute has outstanding core facilities that offer cutting edge instruments and tailored services from expert staff
Latest News & Updates
Find out all our latest news
Careers that have a lasting impact on cancer research and patient care
We are always on the lookout for talented and motivated people to join us. Whether your background is in biological or chemical sciences, mathematics or finance, computer science or logistics, use the links below to see roles across the Institute in our core facilities, operations teams, research groups, and studentships within our exceptional graduate programme.